CellNOpt (from CellNetOptimizer; a.k.a. CNO) is a software used for creating logic-based models of signal transduction networks using different logic formalisms (Boolean, Fuzzy, or differential equations). CellNOpt uses information on signaling pathways encoded as a Prior Knowledge Network, and trains it against high-throughput biochemical data to create cell-specific models.
CellNOpt is freely available under GPL license in R and Matlab languages. It can be also accessed through a python wrapper, and a Cytoscape plugin called CytoCopter provides a graphical user interface.
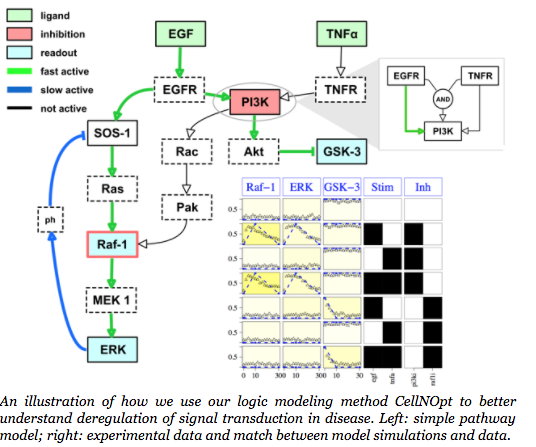
Please use this reference to cite CellNOpt:
E Gjerga, P Trairatphisan, A Gabor, H Koch, C Chevalier, F Ceccarelli, A Dugourd, A Mitsos, J Saez-Rodriguez, Converting networks to predictive logic models from perturbation signalling data with CellNOpt. Bioinformatics, Volume 36, Issue 16, 15 August 2020, Pages 4523–4524,PDF
C Terfve, T Cokelaer, A MacNamara, D Henriques, E Goncalves, MK Morris, M van Iersel, DA Lauffenburger, J Saez-Rodriguez. CellNOptR: a flexible toolkit to train protein signaling networks to data using multiple logic formalisms. BMC Systems Biology, 2012, 6:133 PDF
@article{Gjerga2020cellnoptr,
author = {Gjerga, Enio and Trairatphisan, Panuwat and Gabor, Attila and Koch, Hermann and Chevalier, Celine and Ceccarelli, Franceco and Dugourd, Aurelien and Mitsos, Alexander and Saez-Rodriguez, Julio},
title = "{Converting networks to predictive logic models from perturbation signalling data with CellNOpt}",
journal = {Bioinformatics},
volume = {36},
number = {16},
pages = {4523-4524},
year = {2020},
month = {06},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btaa561},
url = {https://doi.org/10.1093/bioinformatics/btaa561},
eprint = {https://academic.oup.com/bioinformatics/article-pdf/36/16/4523/33965427/btaa561.pdf}
}
We have also developed PHONEMeS, a related tool to build logic models from discovery mass-spectrometry based Phosphoproteomic data. PHONEMeS is described in this paper:
CDA Terfve, E Wilkes, P Casado, P R Cutillas, J Saez-Rodriguez. Large-scale models of signal propagation in human cells derived from discovery phosphoproteomic data. Nature Communications, 2015, 6:8033 PDF. You can also visit PHONEMeS dedicated webpage
A series of packages are available in R. The core CellNOpt is available on BioConductor web site: CellNOptR, revision 1.40.0. The most recent updates from Gjerga, Trairatphisan, Gabor et al. 2020 are available here.
CellNOptR contains the core functions as well as the boolean and steady states version. It implements the workflow described in Saez-Rodriguez et al Mol Sys Bio 2009, with extended capabilities for multiple time points.
CNORdt is an extension that allows to train a Boolean model agains time-courses of data.
CNORfuzzy is an extension to CellNOptR that allows to handle continous values, using constrained fuzzy logic, as described in Morris et al Plos Comp Bio 2011.
CNORprob is a probabilistic logic variant of CellNOpt which allows for quantitative optimisation of logical network for (quasi-)steady-state data as described in Gjerga, Trairatphisan, Gabor et al 2020.
CNORode is an ODE add-on to CellNOptR. It is based on the method of (Wittmann et al BMC Sys Bio 2009), also implemented in the tool Odefy (Krusiek et al BMC Bioinf 2010).
CNORfeeder is an add-on to CellNOptR that permits to extend a network derived from literature with links derived in a strictly data-driven way and supported by protein-protein interactions as described in (Eduati et al Bioinformatics 2012). The most recent version of CNORfeeder, which can also be applied to timecourse data with a logic ordinary differential equations (ODE) formalism can be found here (Gjerga, Trairatphisan, Gabor et al 2020).
CellNOptR-MaBoSS is a method for training boolean logic models of signalling networks using prior knowledge networks and perturbation data with a stochastic simulator (Gjerga, Trairatphisan, Gabor et al 2020).
ShinyCNOR allows using CellNOptR in an interactive way without coding (Gjerga, Trairatphisan, Gabor et al 2020).
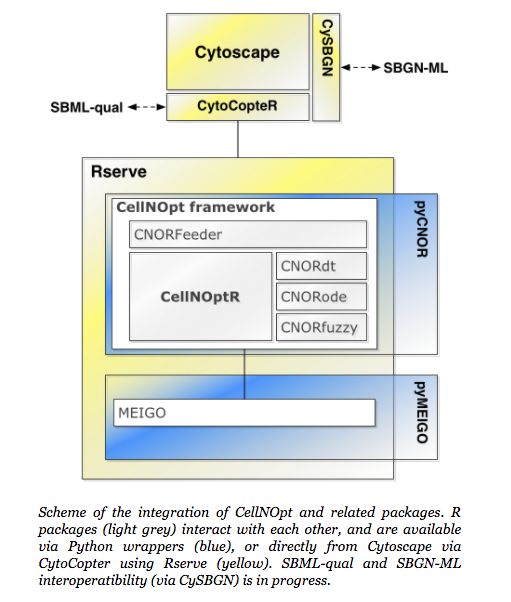
Additional features to the CellNOpt modelling family have been introduced: CellNOpt-ILP, CellNOpt-MaBoSS, CNORprob, Dynamic-Feeder, Post-hoc analysis and the Shiny application (see figure below). These new features have been described in detail in the Gjerga, Trairatphisan, Gabor et al. study.
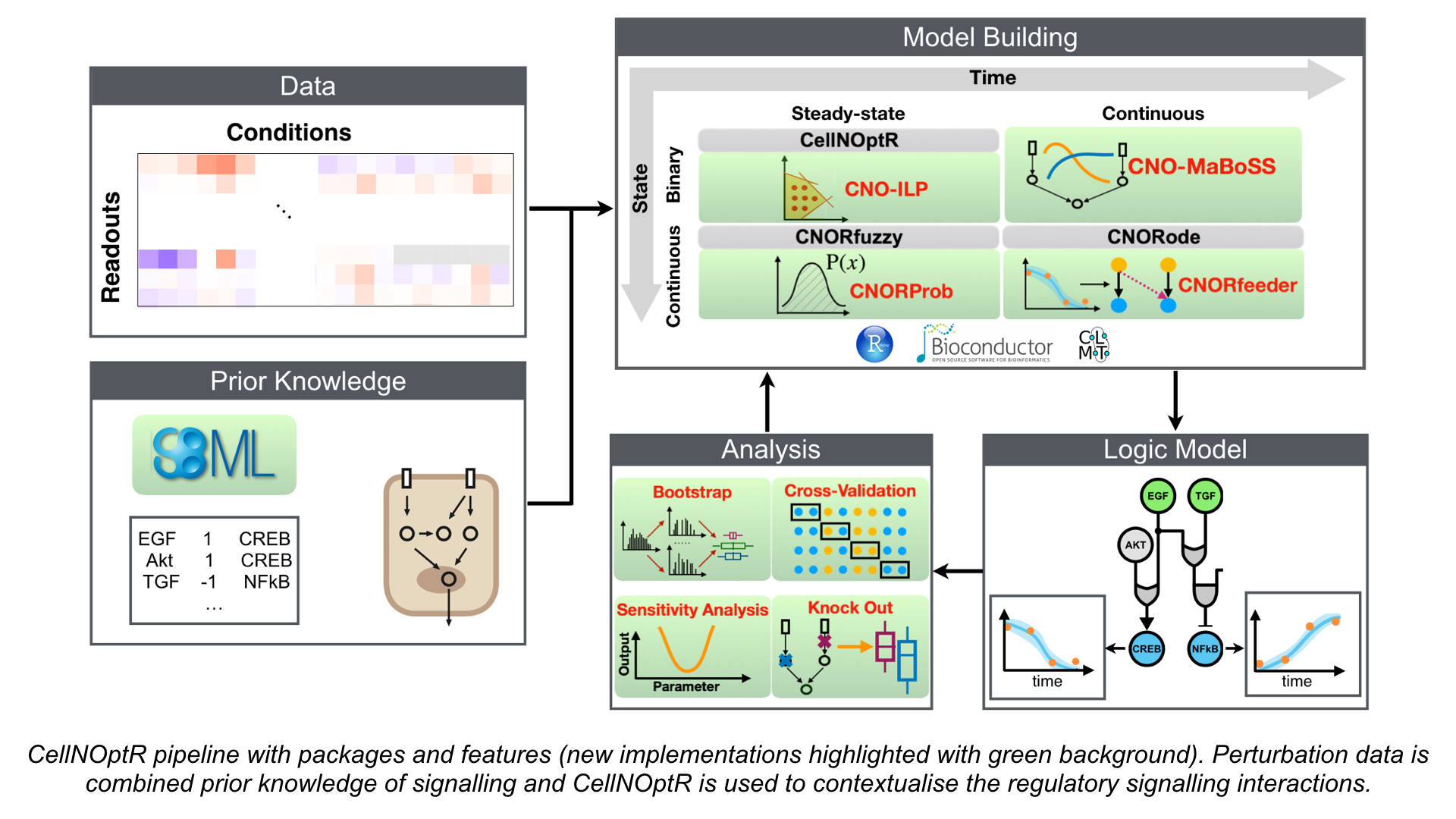
Some features of CellNOpt are also available as a MATLAB toolbox, along with the toolbox Q2LM to analyze models, here
A Python package called cellnopt.wrapper provides a python interace to the R packages (CellNOptR, CNORode and CNORfuzzy). It uses rpy2 and is available on Pypi. For more details see the sphinx documentation in the ./doc directory after downloading the wrapper. In addition a pure Python version is developed on github.
CytoCopteR is a Graphical User Interface designed as a Cytoscape plugin. It provides an interface to CellNOptR using Rserve. More information is available on CytoCopter page.
MEIGO, a global optimization toolbox that includes a number of metaheuristic methods as well as a Bayesian inference method for parameter estimation, that can be applied to model training in CellNOpt. Available in R, Matlab, and Python. Presented in Egea et al BMC Bioinformatics, 214.
Caspo a Python toolbox based on Answer Set Programming to exactly and exhaustively train Boolean models as defined in CellNOpt’s Boolean steady state case. Presented in Guziolowski et al Bioinformatics, 2013 link
The ColoMoTo consortium involves other groups developing tools and methods for logic modelling. We have also jointly develop the standard SBML-qual (Chaouiya et al, BMC Syst Bio 2013) that allows to exchange models within tools.
PHONEMeS Toolbox dedicated to mass spectrometry analysis.
The R packages are self documented. Tutorials and manual are provided on the bioconductor site of each package. Here below are direct links to the Bioconductor vignettes:
Some extra materials and courses about the formats used can be found in the CNODocs. Besides, the following link provides a tutorial given at In Silico Systems Biology, 2013. The following link provides also a CytoCopteR tutorial.
Main reference describing CellNOpt, which can be used to cite it:
E Gjerga, P Trairatphisan, A Gabor, H Koch, C Chevalier, F Ceccarelli, A Dugourd, A Mitsos, J Saez-Rodriguez, Converting networks to predictive logic models from perturbation signalling data with CellNOpt. Bioinformatics, Volume 36, Issue 16, 15 August 2020, Pages 4523–4524.
C Terfve, T Cokelaer, A MacNamara, D Henriques, E Goncalves, MK Morris, M van Iersel, DA Lauffenburger, J Saez-Rodriguez. CellNOptR: a flexible toolkit to train protein signaling networks to data using multiple logic formalisms. BMC Systems Biology, 2012, 6:133 PDF