PHONEMeS (PHOsphorylation NEtworks for Mass Spectrometry) is a method to model signaling networks based on untargeted phosphoproteomics mass spectrometry data and kinase/phosphatase-substrate (K/P-S) interactions. This is a tool currently being developed by the saezlab members. The tool has been implemented as an R-package which contains the accompanying R functions as well as several examples about how to run a PHONEMeS analysis. The method was originally introduced from Terfve et.al. 2015.
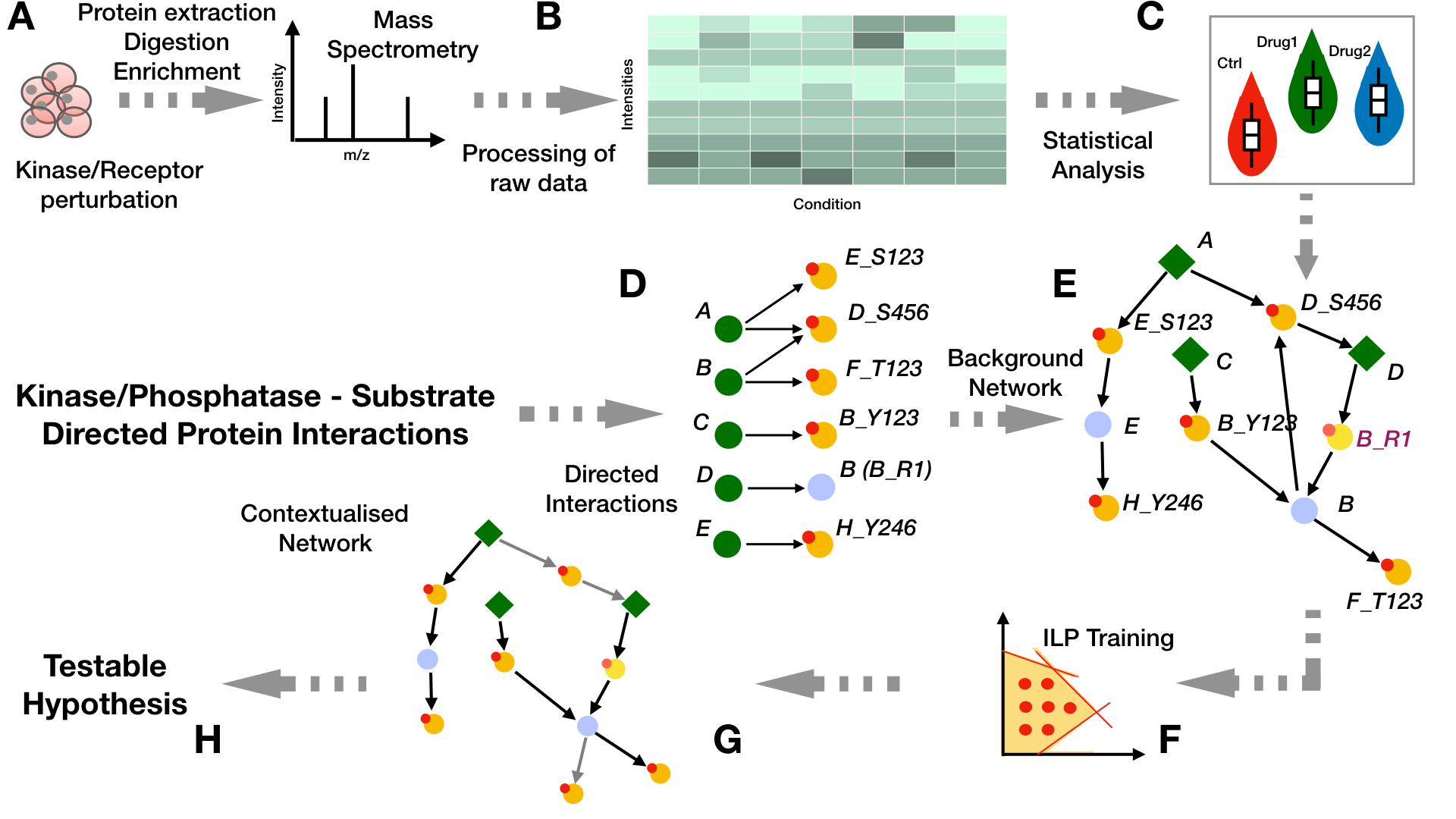
Recently PHONEMeS has been has been reformulated as an Integer Linear Programming (PHONEMeS-ILP, Gjerga et.al. 2021) which is orders of magnitude faster than the original implementation, it requires no cluster infrastructure to run the analysis and expands the scenarios that can be analyzed (Figure 1).
Information about the various PHONEMeS variants implemented and usage details and pipeline can be found here.
If you use any of the ILP implementations of PHONEMeS (PHONEMeS-ILP/PHONEMeS-mult/PHONEMeS-UD), please cite:
Gjerga E., Dugourd A., Tobalina L., Sousa A., Saez-Rodriguez J. (2021). PHONEMeS: Efficient Modeling of Signaling Networks Derived from Large-Scale Mass-Spectrometry Data J Prot Res, 2021, DOI: 10.1021/acs.jproteome.0c00958.
If you use the original implementation of PHONEMeS or if you would like to make mention of the tool, please additionally cite:
Terfve C. D. A., Wilkes E. H., Casado P., Cutillas P. R., and Saez-Rodriguez J. (2015). Large-scale models of signal propagation in human cells derived from discovery phosphoproteomic data. Nature Communications, 6:8033.
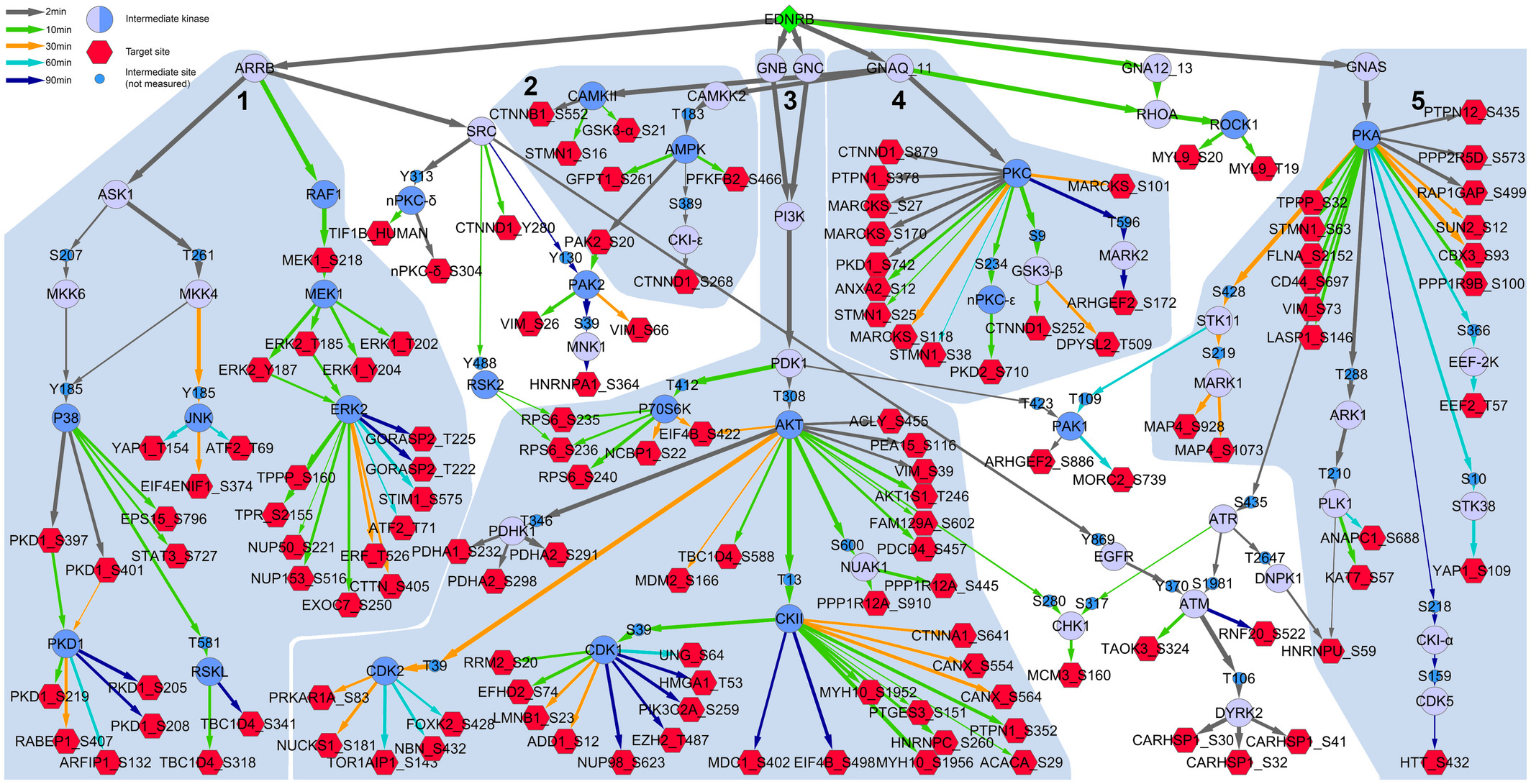
Application of PHONEMeS-ILP over time-resolved data:
Schäfer A., Gjerga E., Welford R.W.D., Renz I., Lehembre F., Groenen P.M.A., Saaez-Rodriguez J., Aebersold R., and Gstaiger M. (August 2019). Elucidating essential kinases of endothelin signalling by logic modelling of phosphoproteomics data Molecular Systems Biology, Volume 15, Issue 8. Codes for this project can be found here.
Application of PHONEMeS-ILP to analyze the effect of hypertensive insults on kidneys:
Rinschen M.M., Palygin O., Guijas C., Palermo A., Palacio-Escat N., Domingo-Almenara X., Montenegro-Burke R., Saez-Rodriguez J., Staruschenko A. and Siuzdak G. (December 2019). Metabolic rewiring of the hypertensive kidney Science Signalling, Vol. 12, Issue 611, eaax9760
Application of PHONEMeS-ILP over colon cancer data:
Gjerga E., Dugourd A., Tobalina L., Sousa A., Saez-Rodriguez J. (2021). PHONEMeS: Efficient Modeling of Signaling Networks Derived from Large-Scale Mass-Spectrometry Data J Prot Res, 2021, DOI: 10.1021/acs.jproteome.0c00958.