CORNETO #
Installation
Get started with CORNETO by following our installation guide for different setups and requirements.
User guide
Learn what CORNETO is and how you can use it to create and modify biological network problems.
Examples & Tutorials
Examples using CORNETO to leverage the power of complex biological networks on real-world data.
API Reference
Python API reference for CORNETO.
Version: 1.0.0.dev5
What is CORNETO?#
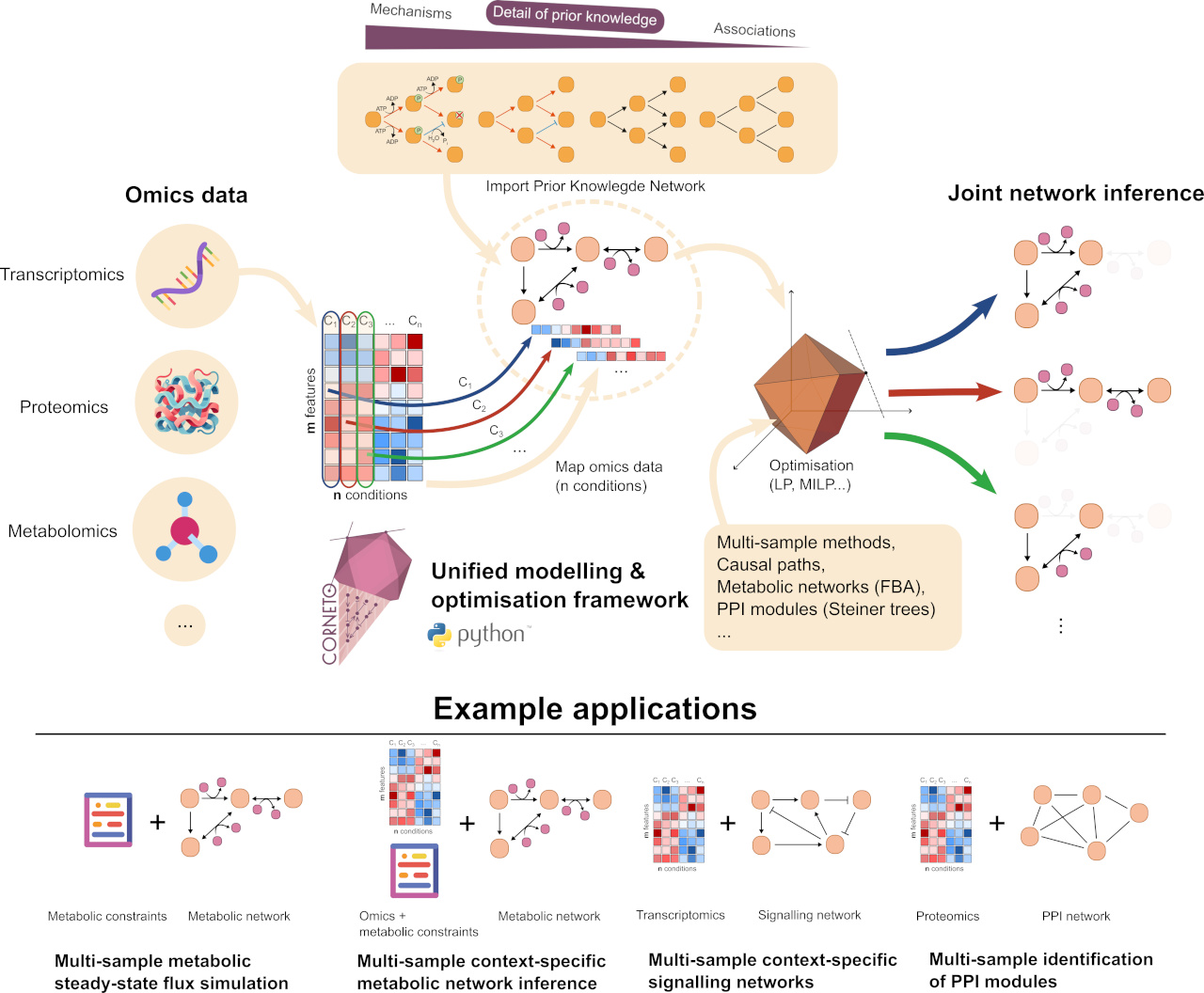
CORNETO (Constraint-based Optimization for the Reconstruction of NETworks from Omics) is a package for unified biological network inference and contextualisation from prior knowledge, developed and maintained by the Saez-Rodriguez Lab. The library is designed with minimal dependencies and is easily extendable, making it a powerful tool for both end-users and developers.
To install CORNETO with open-source mathematical solvers (HIGHs and SCIP), use the following command:
# To install the development version, including the open-source solvers HIGHs and SCIP, use:
!pip install git+https://github.com/saezlab/corneto.git@dev scipy pyscipopt highspy cvxpy
# If you have a license for Gurobi (free for academic use), you can install it with:
pip install gurobipy
Acknowledgements#
CORNETO is developed at the Institute for Computational Biomedicine (Heidelberg University). The development of this project is supported by European Union’s Horizon 2020 Programme under PerMedCoE project (permedcoe.eu) agreement no. 951773 and DECIDER (965193).