transcriptutorial
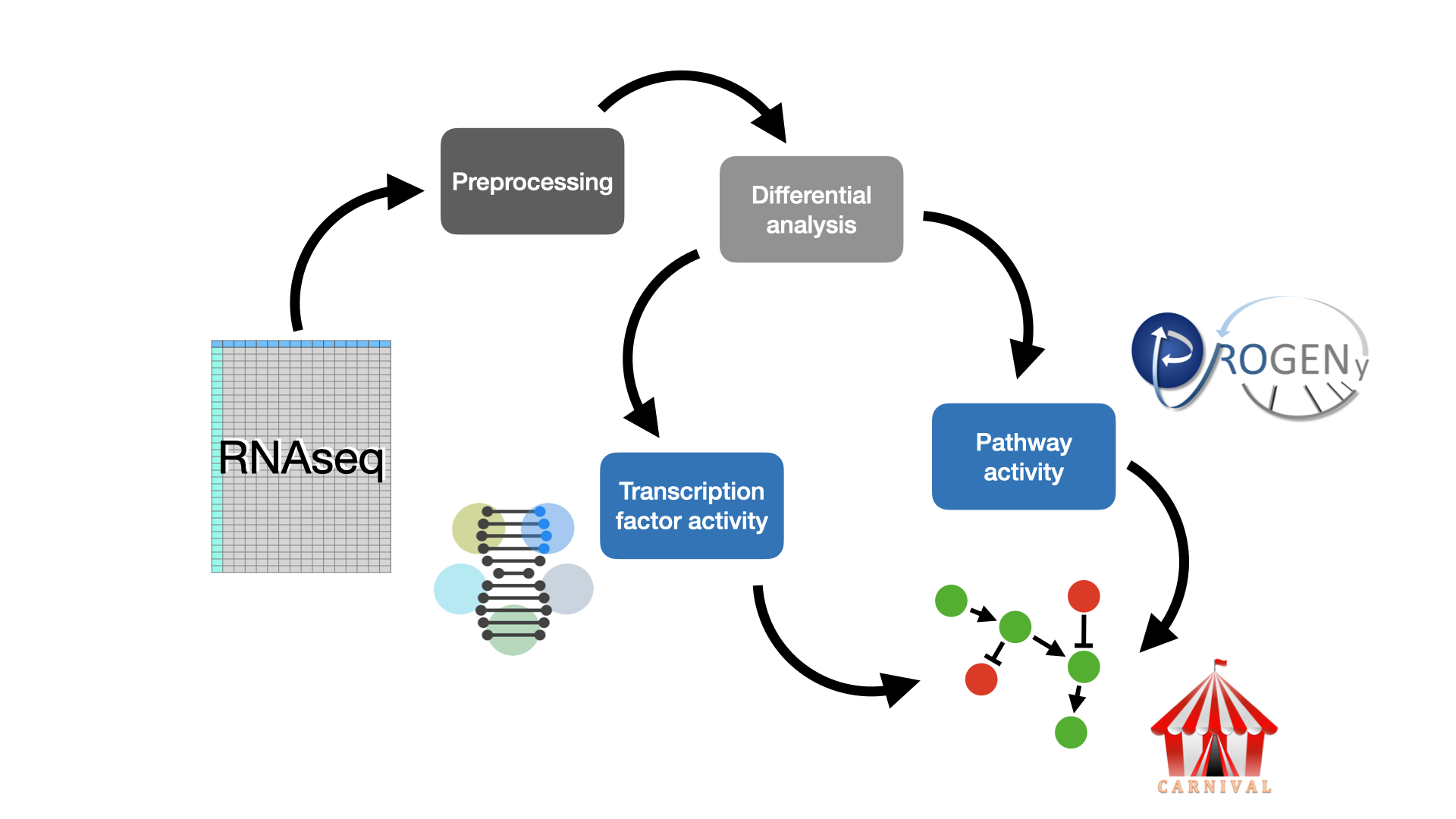
This is a tutorial to guide the analysis of RNAseq datasets using footprint based tools such as decoupleR and CARNIVAL.
For details instructions about decoupleR for RNA data processing and TF activity estiamtion, please refer to: https://saezlab.github.io/decoupleR/articles/tf_bk.html
In script/run_carnival.R:
run_carnival.R uses the CARNIVAL pipeline to analyze gene expression data and identify regulated pathways linking perturbed targets to active transcription factors. It reads in differential activities of transcription factors and a prior knowledge network, applies linear constraints, and uses an ILP solver to minimize fitting error and model size.