LIANA enables the use of any combination of ligand-receptor methods and resources, and their consensus. A faster and memory efficient Python implementation is available here.
Install LIANA
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
if (!requireNamespace("remotes", quietly = TRUE))
install.packages("remotes")
remotes::install_github('saezlab/liana')Tutorial
See a tutorial how to use LIANA to run any combination of 7 methods, plus their consensus, and 16 different resources! The tutorial with the test data takes minutes to complete!
Additional tutorials:
We also refer users to the Cell-cell communication chapter in the best-practices book from Theis lab, as it provides an overview of the common limitations and assumptions in CCC inference from (single-cell) transcriptomics data.
LIANA Framework
The continuous developments of single-cell RNA-Seq (scRNA-Seq) have sparked an immense interest in understanding intercellular crosstalk. Multiple tools and resources that aid the investigation of cell-cell communication (CCC) were published recently. However, these methods and resources are usually in a fixed combination of a tool and its corresponding resource, but in principle any resource could be combined with any method.
To this end, we built a framework to decouple the methods from their corresponding resources.
LIANA also goes a step further as it provides:
A robust and extendable architecture that aims to accelerate method development and benchmarks
A rank aggregate from the results of different methods
A customizable plethora of resources
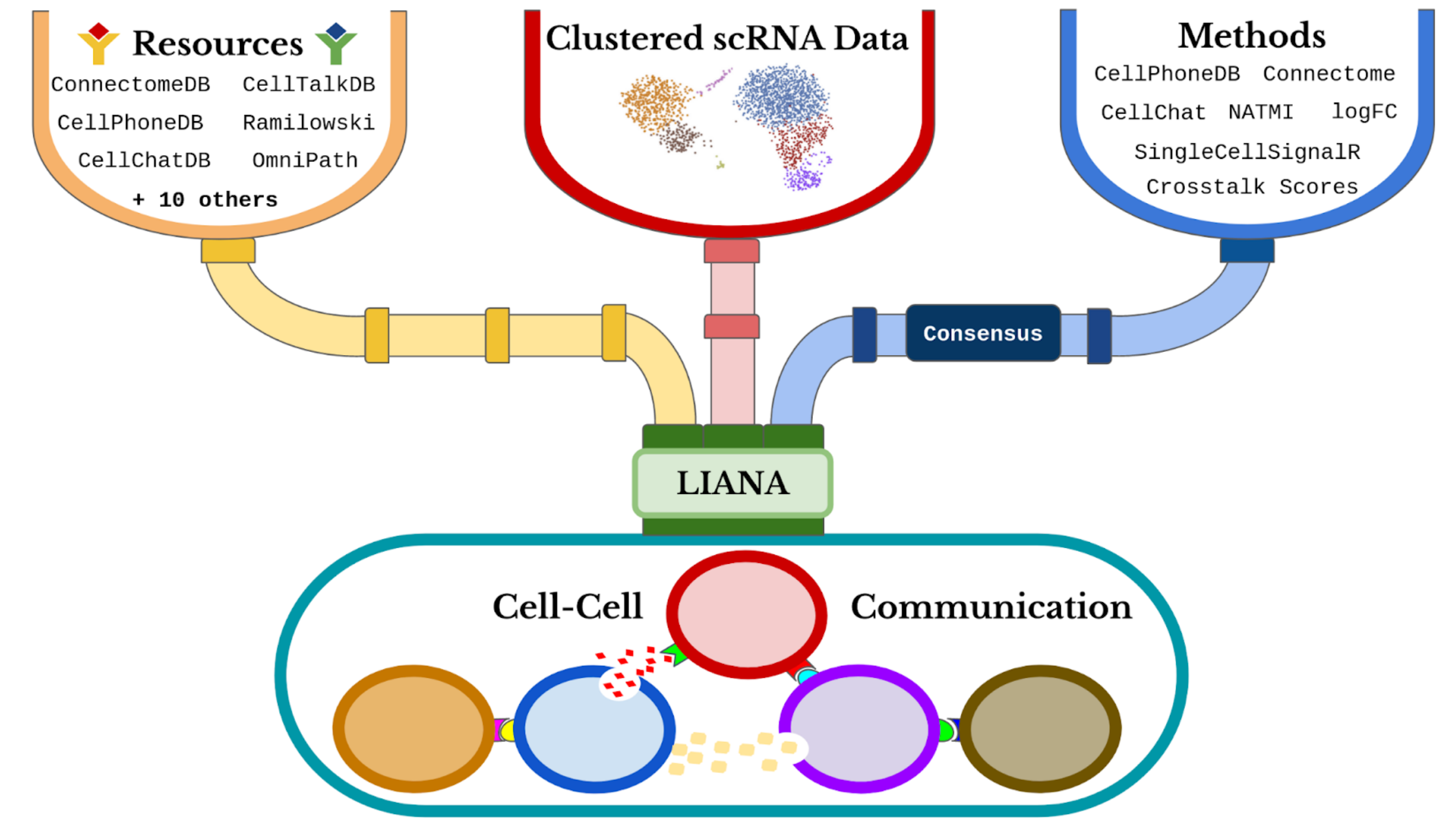
landingpage
Tools
The tools implemented in this repository are:
CellPhoneDBv2 (*, $)
NATMI (*, $)
Connectome (
edge_weights) (*, $)SingleCellSignalR (
LRscores) (SCA) (*, $)iTALK-inspired 1-vs-rest LogFC score (
logfc_comb) (*, $)CytoTalk-inspired
cross-talkscores (*)consensus_rankof the predictions is also provided using the RobustRankAggregate method
*The scoring systems from these methods were re-implemented in LIANA in order to account for multi-meric complexes, simplify the calls to the individual pipelines, or reduce any possible inconsistencies and redundancies in their downstream integration. If you wish to run LIANA with the original tools please see LIANA++.
$ Default methods in LIANA.
Resources
Cell-cell Communication resources
The following CCC resources are accessible via this pipeline:
Consensus
CellCall
CellChatDB
CellPhoneDB
Ramilowski2015
Baccin2019
LRdb
Kiroauc2010
ICELLNET
iTALK
EMBRACE
HPMR
Guide2Pharma
ConnectomeDB2020
CellTalkDB
OmniPath [Deprecated]
Consensus Resource
LIANA’s default resource was generated from the Consensus of several expert-curated resources, then filtered to additional quality control steps including literature support, complex re-union/consensus, and localisation.
OmniPath
All the resources above are retrieved from OmniPath, and more specifically OmnipathR. However, individual resources retrieved from the OmniPath web service are not to be affected by this, as each resource expected to be identical to its original form, apart from minor processing steps.
OmniPath itself serves as a composite CCC resource combining all the ones listed above + more. OmniPath also collects further information about the roles and localisation of proteins in intercellular communication.
We made use of this information to generate the Consensus resource. To obtain more information how we filtered the default Consensus resource, as well as to explore custom filter options see customizing OmniPath resources.
LIANA++
If you are interested in making use of the LIANA architecture for your own method, this vignette provides instructions how to obtain a comprehensive table of LR statistics, which can then be used by custom scoring functions. In the same vignette are also instructions how to install and run the original methods via a convenient R wrapper, e.g. for their benchmark.
Contact
We appreciate any feedback, so please do not hesitate to open an issue on the liana github page!
NEWS
We are commited to the further development of LIANA and we refer the users to the NEWS page!
Citing LIANA:
Dimitrov, D., Türei, D., Garrido-Rodriguez M., Burmedi P.L., Nagai, J.S., Boys, C., Flores, R.O.R., Kim, H., Szalai, B., Costa, I.G., Valdeolivas, A., Dugourd, A. and Saez-Rodriguez, J. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat Commun 13, 3224 (2022). https://doi.org/10.1038/s41467-022-30755-0
Also, if you use the OmniPath CCC Resource for your analysis, please cite:
Türei, D., Valdeolivas, A., Gul, L., Palacio‐Escat, N., Klein, M., Ivanova, O., Ölbei, M., Gábor, A., Theis, F., Módos, D. and Korcsmáros, T., 2021. Integrated intra‐and intercellular signaling knowledge for multicellular omics analysis. Molecular systems biology, 17(3), p.e9923. https://doi.org/10.15252/msb.20209923
