Chronic CCl4+EtOH mouse model
Last updated: 2021-07-06
Checks: 7 0
Knit directory: liver-disease-atlas/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20201218) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 69f7892. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/01-mouse-chronic-ccl4_cache/
Ignored: analysis/01.2-mouse-chronic-ccl4-wtd_cache/
Ignored: analysis/02-mouse-acute-apap_cache/
Ignored: analysis/03-mouse-acute-ccl4_cache/
Ignored: analysis/04-mouse-acute-ph_cache/
Ignored: analysis/05-mouse-acute-bdl_cache/
Ignored: analysis/06-mouse-acute-lps_cache/
Ignored: analysis/07-mouse-acute-tunicamycin_cache/
Ignored: analysis/08-human-diehl-nafld_cache/
Ignored: analysis/09-human-hampe13-nash_cache/
Ignored: analysis/10-human-hampe14-misc_cache/
Ignored: analysis/11-human-hoang-nafld_cache/
Ignored: analysis/12-human-ramnath-fibrosis_cache/
Ignored: analysis/13-meta-chronic-vs-acute_cache/
Ignored: analysis/14-meta-mouse-vs-human_cache/
Ignored: analysis/15-plot-chronic-ccl4_cache/
Ignored: analysis/16-plot-acute-apap_cache/
Ignored: analysis/17-plot-acute-ccl4_cache/
Ignored: analysis/18-plot-acute-ph_cache/
Ignored: analysis/19-plot-acute-bdl_cache/
Ignored: analysis/20-plot-study-overview_cache/
Ignored: analysis/21-plot-chronic-vs-acute_cache/
Ignored: analysis/22-plot-mouse-vs-human_cache/
Ignored: analysis/23-plot-precision-recall_cache/
Ignored: analysis/24-save-tables_cache/
Ignored: code/.DS_Store
Ignored: code/README.html
Ignored: code/meta-mouse-vs-human/.DS_Store
Ignored: data.zip
Ignored: data/.DS_Store
Ignored: data/Icon
Ignored: data/annotation/
Ignored: data/human-diehl-nafld/
Ignored: data/human-hampe13-nash/
Ignored: data/human-hampe14-misc/
Ignored: data/human-hoang-nafld/
Ignored: data/human-ramnath-fibrosis/
Ignored: data/meta-chronic-vs-acute/
Ignored: data/meta-mouse-vs-human/
Ignored: data/mouse-acute-apap/
Ignored: data/mouse-acute-bdl/
Ignored: data/mouse-acute-ccl4/
Ignored: data/mouse-acute-lps/
Ignored: data/mouse-acute-ph/
Ignored: data/mouse-acute-tunicamycin/
Ignored: data/mouse-chronic-ccl4-wtd/
Ignored: data/mouse-chronic-ccl4/
Ignored: external_software/.DS_Store
Ignored: external_software/README.html
Ignored: external_software/stem/.DS_Store
Ignored: figures/.DS_Store
Ignored: figures/Figure 1 (partial).pdf
Ignored: figures/Figure 1.pdf
Ignored: figures/Figure 1.png
Ignored: figures/Figure 2 (partial).pdf
Ignored: figures/Figure 2.pdf
Ignored: figures/Figure 2.png
Ignored: figures/Figure 3.pdf
Ignored: figures/Figure 3.png
Ignored: figures/Figure 4.pdf
Ignored: figures/Figure 4.png
Ignored: figures/Figure 5.pdf
Ignored: figures/Figure 5.png
Ignored: figures/Figure 6.pdf
Ignored: figures/Figure 6.png
Ignored: figures/Icon
Ignored: figures/Supplementary Figure 0.1.pdf
Ignored: figures/Supplementary Figure 0.1.png
Ignored: figures/Supplementary Figure 1.1.pdf
Ignored: figures/Supplementary Figure 1.1.png
Ignored: figures/Supplementary Figure 2.1.pdf
Ignored: figures/Supplementary Figure 2.1.png
Ignored: figures/Supplementary Figure 2.2.pdf
Ignored: figures/Supplementary Figure 2.2.png
Ignored: figures/Supplementary Figure 2.3.pdf
Ignored: figures/Supplementary Figure 2.3.png
Ignored: figures/Supplementary Figure 2.4.pdf
Ignored: figures/Supplementary Figure 2.4.png
Ignored: figures/Supplementary Figure 2.5.pdf
Ignored: figures/Supplementary Figure 2.5.png
Ignored: figures/Supplementary Figure 2.6.pdf
Ignored: figures/Supplementary Figure 2.6.png
Ignored: figures/Supplementary Figure 2.7.pdf
Ignored: figures/Supplementary Figure 2.7.png
Ignored: figures/Supplementary Figure 3.1.pdf
Ignored: figures/Supplementary Figure 3.1.png
Ignored: figures/Supplementary Figure 3.2.pdf
Ignored: figures/Supplementary Figure 3.2.png
Ignored: figures/Supplementary Figure 3.3.pdf
Ignored: figures/Supplementary Figure 3.3.png
Ignored: figures/Supplementary Figure 3.4.pdf
Ignored: figures/Supplementary Figure 3.4.png
Ignored: figures/Supplementary Figure 4.1.pdf
Ignored: figures/Supplementary Figure 4.1.png
Ignored: figures/Supplementary Figure 4.2.pdf
Ignored: figures/Supplementary Figure 4.2.png
Ignored: figures/Supplementary Figure 6.1.pdf
Ignored: figures/Supplementary Figure 6.1.png
Ignored: figures/Supplementary Figure 6.2.pdf
Ignored: figures/Supplementary Figure 6.2.png
Ignored: figures/figures.key
Ignored: figures/histologies.key
Ignored: figures/panels/
Ignored: figures/tmp/.DS_Store
Ignored: figures/tmp/Fig5A1.pdf
Ignored: figures/tmp/Fig5A2.pdf
Ignored: figures/tmp/Icon
Ignored: geo_submission/
Ignored: output/.DS_Store
Ignored: output/Icon
Ignored: output/README.html
Ignored: output/human-diehl-nafld/Icon
Ignored: output/human-diehl-nafld/limma_result.rds
Ignored: output/human-diehl-nafld/meta_data.rds
Ignored: output/human-diehl-nafld/normalized_expression.rds
Ignored: output/human-diehl-nafld/pca_result.rds
Ignored: output/human-diehl-nafld/z_scores.rds
Ignored: output/human-hampe13-nash/Icon
Ignored: output/human-hampe13-nash/limma_result.rds
Ignored: output/human-hampe13-nash/meta_data.rds
Ignored: output/human-hampe13-nash/normalized_expression.rds
Ignored: output/human-hampe13-nash/pca_result.rds
Ignored: output/human-hampe13-nash/z_scores.rds
Ignored: output/human-hampe14-misc/Icon
Ignored: output/human-hampe14-misc/limma_result.rds
Ignored: output/human-hampe14-misc/meta_data.rds
Ignored: output/human-hampe14-misc/normalized_expression.rds
Ignored: output/human-hampe14-misc/pca_result.rds
Ignored: output/human-hampe14-misc/z_scores.rds
Ignored: output/human-hoang-nafld/Icon
Ignored: output/human-hoang-nafld/limma_result.rds
Ignored: output/human-hoang-nafld/normalized_expression.rds
Ignored: output/human-hoang-nafld/pca_result.rds
Ignored: output/human-hoang-nafld/z_scores.rds
Ignored: output/human-ramnath-fibrosis/Icon
Ignored: output/human-ramnath-fibrosis/limma_result.rds
Ignored: output/human-ramnath-fibrosis/normalized_expression.rds
Ignored: output/human-ramnath-fibrosis/pca_result.rds
Ignored: output/human-ramnath-fibrosis/z_scores.rds
Ignored: output/meta-chronic-vs-acute/Icon
Ignored: output/meta-chronic-vs-acute/acute_gene_pool.rds
Ignored: output/meta-chronic-vs-acute/chronic_gene_pool.rds
Ignored: output/meta-chronic-vs-acute/exclusive_genes_characterization.rds
Ignored: output/meta-chronic-vs-acute/gene_membership.rds
Ignored: output/meta-chronic-vs-acute/gene_set_similarity.rds
Ignored: output/meta-chronic-vs-acute/go_cluster_ranking.rds
Ignored: output/meta-chronic-vs-acute/go_wordcounts.rds
Ignored: output/meta-chronic-vs-acute/interstudy_enrichment.rds
Ignored: output/meta-chronic-vs-acute/limma_result.rds
Ignored: output/meta-chronic-vs-acute/meta_data.rds
Ignored: output/meta-chronic-vs-acute/pca_dist.rds
Ignored: output/meta-chronic-vs-acute/ranked_common_genes.rds
Ignored: output/meta-chronic-vs-acute/ranked_exclusive_acute_genes.rds
Ignored: output/meta-chronic-vs-acute/ranked_exclusive_chronic_genes.rds
Ignored: output/meta-chronic-vs-acute/union_acute_geneset.rds
Ignored: output/meta-chronic-vs-acute/union_chronic_geneset.rds
Ignored: output/meta-chronic-vs-acute/z_score_pca.rds
Ignored: output/meta-mouse-vs-human/Icon
Ignored: output/meta-mouse-vs-human/basel_gene_expression_levels.rds
Ignored: output/meta-mouse-vs-human/chronic_mouse_deg_numbers.rds
Ignored: output/meta-mouse-vs-human/consistent_genes.rds
Ignored: output/meta-mouse-vs-human/cross_species_enrichment.rds
Ignored: output/meta-mouse-vs-human/cross_species_similarity.rds
Ignored: output/meta-mouse-vs-human/etiology_gene_sets.rds
Ignored: output/meta-mouse-vs-human/gene_set_similarity.rds
Ignored: output/meta-mouse-vs-human/go_cluster_ranking.rds
Ignored: output/meta-mouse-vs-human/go_wordcounts.rds
Ignored: output/meta-mouse-vs-human/gsea_res.rds
Ignored: output/meta-mouse-vs-human/individual_le.rds
Ignored: output/meta-mouse-vs-human/interstudy_enrichment.rds
Ignored: output/meta-mouse-vs-human/leading_edges.rds
Ignored: output/meta-mouse-vs-human/leading_edges_characterization.rds
Ignored: output/meta-mouse-vs-human/leading_edges_mgi.rds
Ignored: output/meta-mouse-vs-human/limma_result.rds
Ignored: output/meta-mouse-vs-human/meta_data.rds
Ignored: output/meta-mouse-vs-human/precision_recall.rds
Ignored: output/meta-mouse-vs-human/precision_recall_chronicity.rds
Ignored: output/meta-mouse-vs-human/teufel_genes.rds
Ignored: output/meta-mouse-vs-human/teufel_genes_hs.rds
Ignored: output/meta-mouse-vs-human/z_score_pca.rds
Ignored: output/mouse-acute-apap/.DS_Store
Ignored: output/mouse-acute-apap/Icon
Ignored: output/mouse-acute-apap/limma_result.rds
Ignored: output/mouse-acute-apap/meta_data.rds
Ignored: output/mouse-acute-apap/normalized_expression.rds
Ignored: output/mouse-acute-apap/pca_result.rds
Ignored: output/mouse-acute-apap/stem/.DS_Store
Ignored: output/mouse-acute-apap/stem/Icon
Ignored: output/mouse-acute-apap/stem/input/Icon
Ignored: output/mouse-acute-apap/stem/input/apap.txt
Ignored: output/mouse-acute-apap/stem_characterization.rds
Ignored: output/mouse-acute-apap/stem_result.rds
Ignored: output/mouse-acute-apap/z_scores.rds
Ignored: output/mouse-acute-bdl/.DS_Store
Ignored: output/mouse-acute-bdl/Icon
Ignored: output/mouse-acute-bdl/limma_result.rds
Ignored: output/mouse-acute-bdl/meta_data.rds
Ignored: output/mouse-acute-bdl/normalized_expression.rds
Ignored: output/mouse-acute-bdl/pca_result.rds
Ignored: output/mouse-acute-bdl/stem/.DS_Store
Ignored: output/mouse-acute-bdl/stem/Icon
Ignored: output/mouse-acute-bdl/stem/input/Icon
Ignored: output/mouse-acute-bdl/stem/input/bdl.txt
Ignored: output/mouse-acute-bdl/stem_characterization.rds
Ignored: output/mouse-acute-bdl/stem_result.rds
Ignored: output/mouse-acute-bdl/z_scores.rds
Ignored: output/mouse-acute-ccl4/.DS_Store
Ignored: output/mouse-acute-ccl4/Icon
Ignored: output/mouse-acute-ccl4/limma_result.rds
Ignored: output/mouse-acute-ccl4/meta_data.rds
Ignored: output/mouse-acute-ccl4/normalized_expression.rds
Ignored: output/mouse-acute-ccl4/pca_result.rds
Ignored: output/mouse-acute-ccl4/stem/.DS_Store
Ignored: output/mouse-acute-ccl4/stem/Icon
Ignored: output/mouse-acute-ccl4/stem/input/Icon
Ignored: output/mouse-acute-ccl4/stem/input/ccl4.txt
Ignored: output/mouse-acute-ccl4/stem_characterization.rds
Ignored: output/mouse-acute-ccl4/stem_result.rds
Ignored: output/mouse-acute-ccl4/z_scores.rds
Ignored: output/mouse-acute-lps/Icon
Ignored: output/mouse-acute-lps/limma_result.rds
Ignored: output/mouse-acute-lps/meta_data.rds
Ignored: output/mouse-acute-lps/normalized_expression.rds
Ignored: output/mouse-acute-lps/pca_result.rds
Ignored: output/mouse-acute-lps/z_scores.rds
Ignored: output/mouse-acute-ph/.DS_Store
Ignored: output/mouse-acute-ph/Icon
Ignored: output/mouse-acute-ph/limma_result.rds
Ignored: output/mouse-acute-ph/meta_data.rds
Ignored: output/mouse-acute-ph/normalized_expression.rds
Ignored: output/mouse-acute-ph/pca_result.rds
Ignored: output/mouse-acute-ph/stem/.DS_Store
Ignored: output/mouse-acute-ph/stem/Icon
Ignored: output/mouse-acute-ph/stem/input/Icon
Ignored: output/mouse-acute-ph/stem/input/hepatec.txt
Ignored: output/mouse-acute-ph/stem_characterization.rds
Ignored: output/mouse-acute-ph/stem_result.rds
Ignored: output/mouse-acute-ph/z_scores.rds
Ignored: output/mouse-acute-tunicamycin/Icon
Ignored: output/mouse-acute-tunicamycin/limma_result.rds
Ignored: output/mouse-acute-tunicamycin/meta_data.rds
Ignored: output/mouse-acute-tunicamycin/normalized_expression.rds
Ignored: output/mouse-acute-tunicamycin/pca_result.rds
Ignored: output/mouse-acute-tunicamycin/z_scores.rds
Ignored: output/mouse-chronic-ccl4/.DS_Store
Ignored: output/mouse-chronic-ccl4/Icon
Ignored: output/mouse-chronic-ccl4/limma_result.rds
Ignored: output/mouse-chronic-ccl4/limma_result_hs.rds
Ignored: output/mouse-chronic-ccl4/normalized_expression.rds
Ignored: output/mouse-chronic-ccl4/pca_result.rds
Ignored: output/mouse-chronic-ccl4/stem/.DS_Store
Ignored: output/mouse-chronic-ccl4/stem/Icon
Ignored: output/mouse-chronic-ccl4/stem/input/Icon
Ignored: output/mouse-chronic-ccl4/stem/input/pure_ccl4.txt
Ignored: output/mouse-chronic-ccl4/stem_characterization.rds
Ignored: output/mouse-chronic-ccl4/stem_result.rds
Ignored: output/mouse-chronic-ccl4/z_scores.rds
Ignored: renv/library/
Ignored: renv/staging/
Ignored: tables/Supplementary Table 1.xlsx
Ignored: tables/Supplementary Table xy consistent_genes.xlsx
Ignored: tables/Supplementary Table xy exclusive_common_genes.xlsx
Ignored: tables/Supplementary Table xy human_degs.xlsx
Ignored: tables/Supplementary Table xy stem_results.xlsx
Untracked files:
Untracked: code/mouse-chronic-ccl4-etoh/
Untracked: code/mouse-chronic-ccl4-wtd/
Untracked: data/mouse-chronic-ccl4-etoh/
Untracked: output/mouse-chronic-ccl4-etoh/
Untracked: output/mouse-chronic-ccl4-wtd/
Unstaged changes:
Modified: README.Rmd
Modified: README.md
Modified: analysis/14-meta-mouse-vs-human.Rmd
Modified: analysis/23-plot-precision-recall.Rmd
Modified: analysis/_site.yml
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/01.1-mouse-chronic-ccl4-etoh.Rmd) and HTML (docs/01.1-mouse-chronic-ccl4-etoh.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 69f7892 | christianholland | 2021-07-06 | analysis of two additional chronic mouse models |
Introduction
Here we analysis a mouse model of CCl4 in combination with alcohol induced chronic liver disease.
Libraries and sources
These libraries and sources are used for this analysis.
library(tidyverse)
library(tidylog)
library(here)
library(edgeR)
library(biobroom)
library(progeny)
library(dorothea)
library(janitor)
library(msigdf) # remotes::install_github("ToledoEM/msigdf@v7.1")
library(AachenColorPalette)
library(cowplot)
library(lemon)
library(patchwork)
library(VennDiagram)
library(gridExtra)
library(ggpubr)
options("tidylog.display" = list(print))
source(here("code/utils-rnaseq.R"))
source(here("code/utils-utils.R"))
source(here("code/utils-plots.R"))Definition of global variables that are used throughout this analysis.
# i/o
data_path <- "data/mouse-chronic-ccl4-etoh"
output_path <- "output/mouse-chronic-ccl4-etoh"
# graphical parameters
# fontsize
fz <- 9Preliminary exploratory analysis
Library size
Barplot of the library size (total counts) for each of the samples.
count_matrix <- readRDS(here(data_path, "count_matrix.rds"))
plot_libsize(count_matrix) +
my_theme(fsize = fz)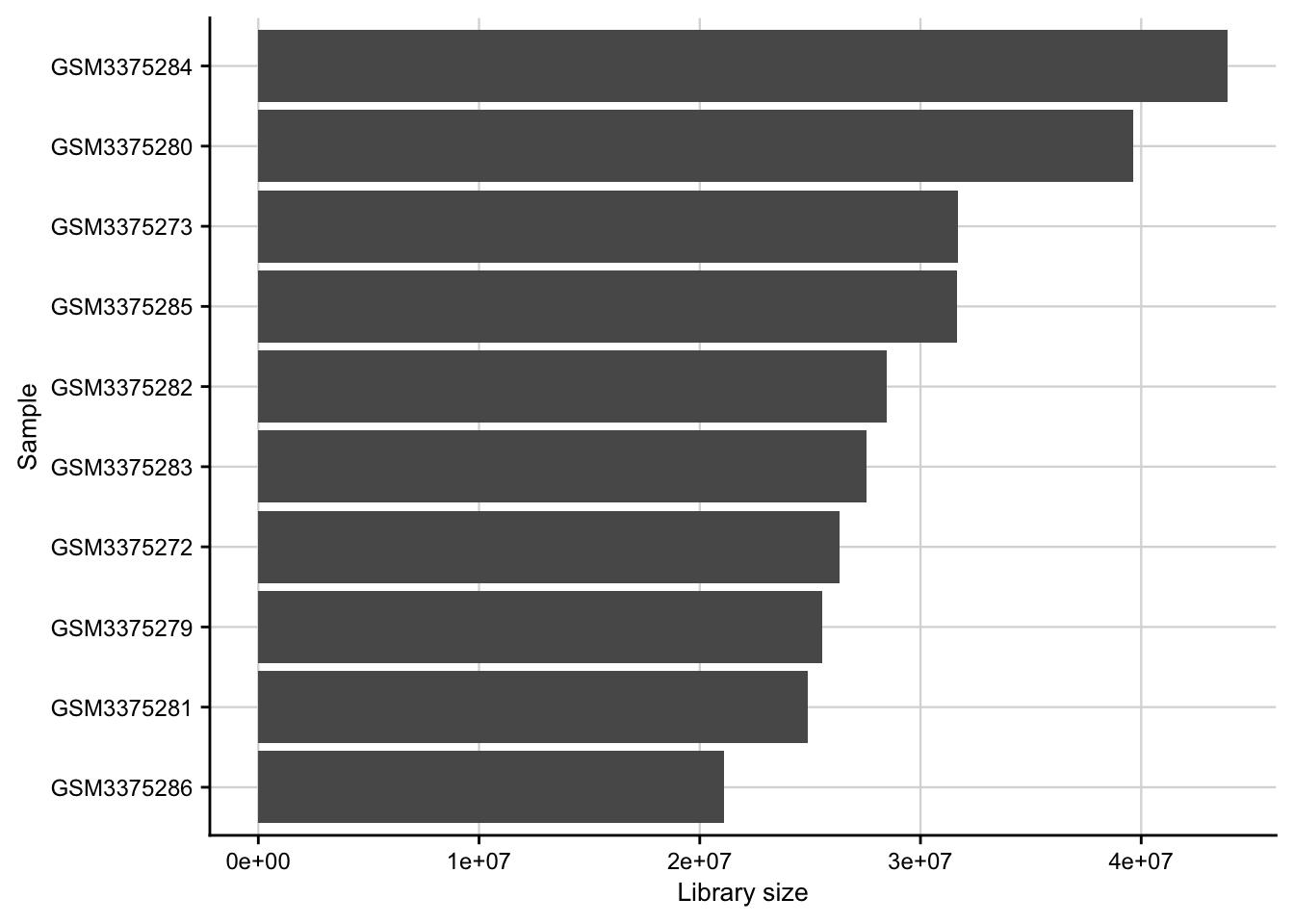
Count distribution
Violin plots of the raw read counts for each of the samples.
count_matrix <- readRDS(here(data_path, "count_matrix.rds"))
meta <- readRDS(here(data_path, "meta_data.rds"))
count_matrix %>%
tdy("gene", "sample", "count", meta) %>%
arrange(treatment) %>%
ggplot(aes(
x = fct_reorder(sample, as.numeric(treatment)), y = log10(count + 1),
group = sample, fill = treatment
)) +
geom_violin() +
theme(
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1),
legend.position = "top"
) +
labs(x = NULL) +
my_theme(grid = "no", fsize = fz)
#> gather: reorganized (GSM3375272, GSM3375273, GSM3375279, GSM3375280, GSM3375281, …) into (sample, count) [was 24504x11, now 245040x3]
#> left_join: added 3 columns (time, treatment, group)
#> > rows only in x 0
#> > rows only in y ( 0)
#> > matched rows 245,040
#> > =========
#> > rows total 245,040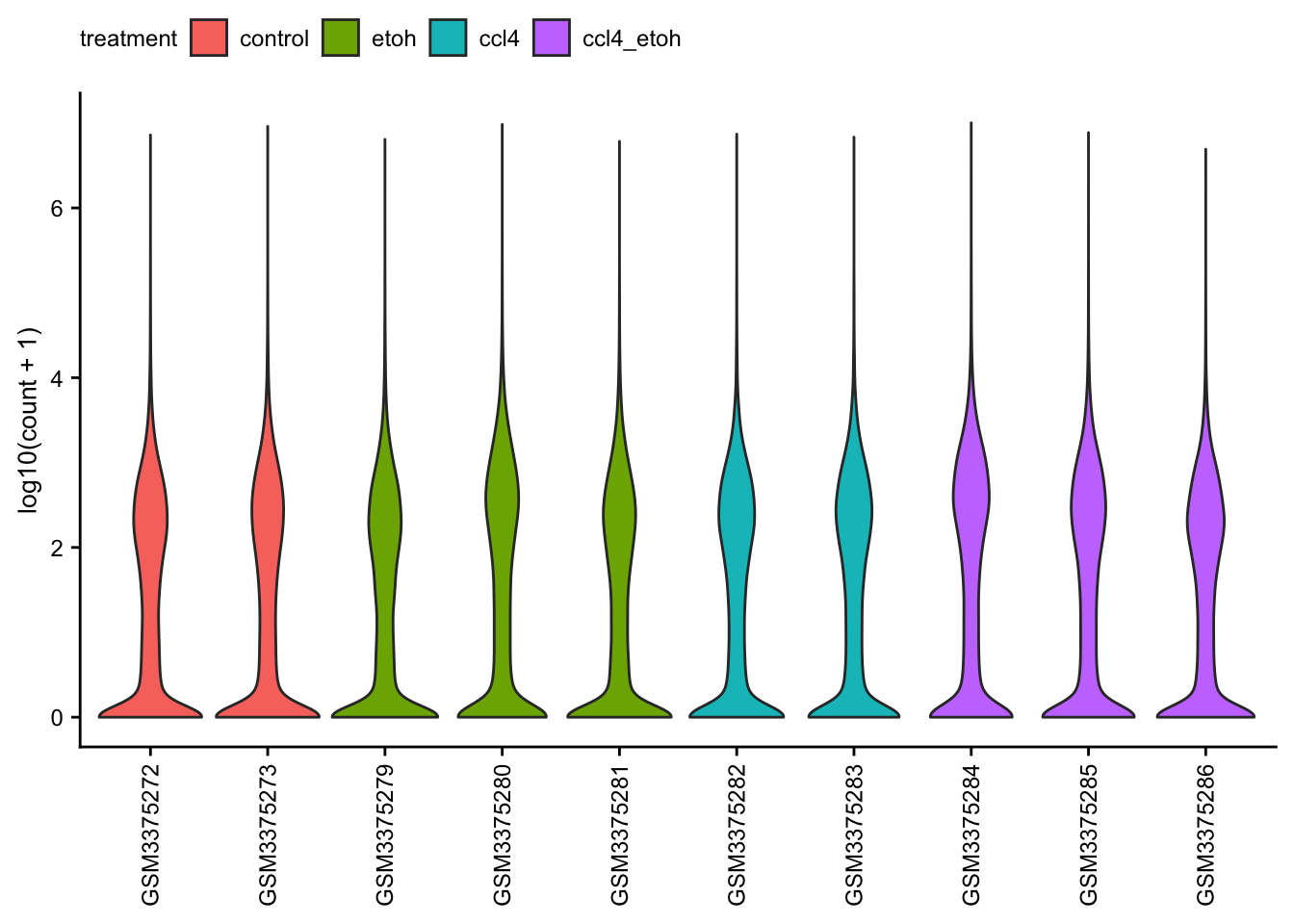
PCA of raw data
PCA plot of raw read counts contextualized based on the time point and treatment. Before gene with a constant expression across all samples are removed and count values are transformed to log2 scale. Only the top 1000 most variable genes are used as features.
count_matrix <- readRDS(here(data_path, "count_matrix.rds"))
meta <- readRDS(here(data_path, "meta_data.rds"))
stopifnot(colnames(count_matrix) == meta$sample)
# remove constant expressed genes and transform to log2 scale
preprocessed_count_matrix <- preprocess_count_matrix(count_matrix)
#> Discarding 5120 genes
#> Keeping 19384 genes
pca_result <- do_pca(preprocessed_count_matrix, meta, top_n_var_genes = 1000)
#> left_join: added 3 columns (time, treatment, group)
#> > rows only in x 0
#> > rows only in y ( 0)
#> > matched rows 10
#> > ====
#> > rows total 10
plot_pca(pca_result, feature = "treatment") &
my_theme(fsize = fz)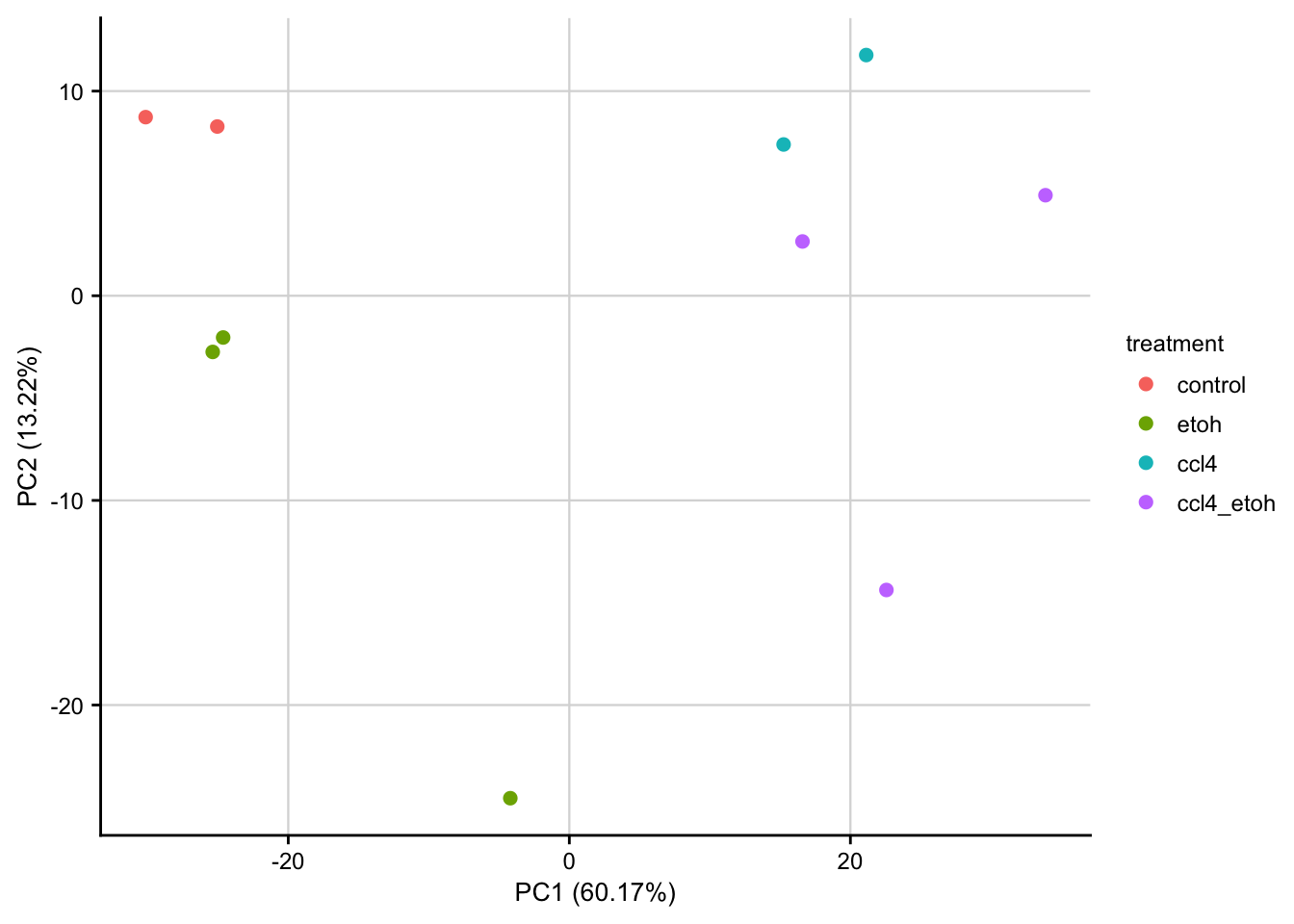
Data processing
Normalization
Raw read counts are normalized by first filtering out lowly expressed genes, TMM normalization and finally logCPM transformation.
count_matrix <- readRDS(here(data_path, "count_matrix.rds"))
meta <- readRDS(here(data_path, "meta_data.rds"))
stopifnot(meta$sample == colnames(count_matrix))
dge_obj <- DGEList(count_matrix, group = meta$group)
# filter low read counts, TMM normalization and logCPM transformation
norm <- voom_normalization(dge_obj)
#> Discarding 10325 genes
#> Keeping 14179 genes
saveRDS(norm, here(output_path, "normalized_expression.rds"))PCA of normalized data
PCA plot of normalized expression data contextualized based on the time point and treatment. Only the top 1000 most variable genes are used as features.
expr <- readRDS(here(output_path, "normalized_expression.rds"))
meta <- readRDS(here(data_path, "meta_data.rds"))
pca_result <- do_pca(expr, meta, top_n_var_genes = 1000)
#> left_join: added 3 columns (time, treatment, group)
#> > rows only in x 0
#> > rows only in y ( 0)
#> > matched rows 10
#> > ====
#> > rows total 10
saveRDS(pca_result, here(output_path, "pca_result.rds"))
plot_pca(pca_result, feature = "treatment") &
my_theme(fsize = fz)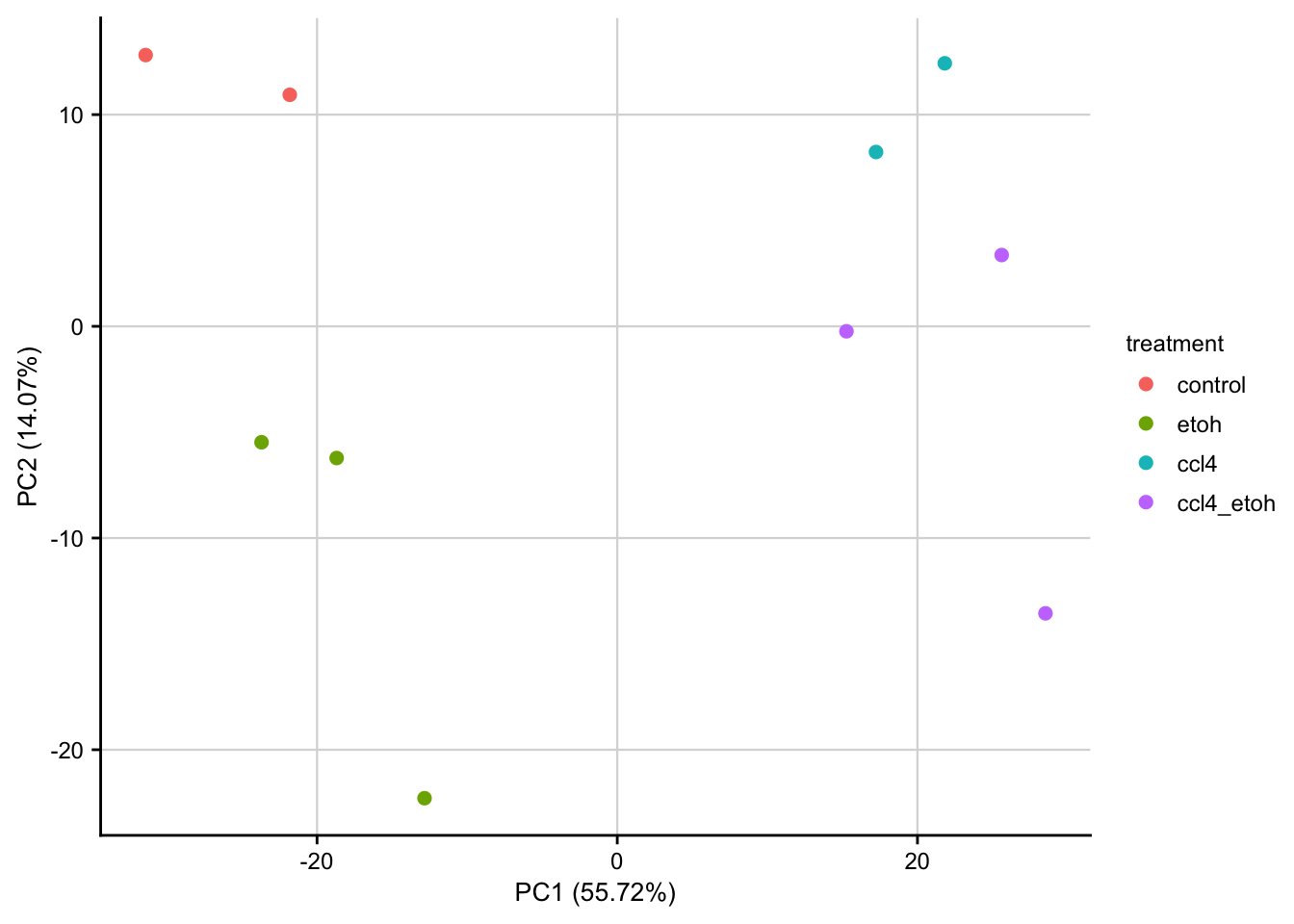
Differential gene expression analysis
### Running limma Differential gene expression analysis via limma with the aim to identify the effect of CCl4 intoxication while regression out the effect of the oil.
# load expression and meta data
expr <- readRDS(here(output_path, "normalized_expression.rds"))
meta <- readRDS(here(data_path, "meta_data.rds"))
stopifnot(colnames(expr) == meta$sample)
# build design matrix
design <- model.matrix(~ 0 + group, data = meta)
rownames(design) <- meta$sample
colnames(design) <- levels(meta$group)
# define contrasts
contrasts <- makeContrasts(
# effect of olive oil
ccl_effect = ccl4_20 - control_0,
etoh_effect = etoh_20 - control_0,
combined_effect = ccl4_etoh_20 - control_0,
levels = design
)
limma_result <- run_limma(expr, design, contrasts) %>%
assign_deg()
#> Warning: `tbl_df()` is deprecated as of dplyr 1.0.0.
#> Please use `tibble::as_tibble()` instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_warnings()` to see where this warning was generated.
#> select: renamed 3 variables (contrast, logFC, pval) and dropped one variable
#> ungroup: no grouping variables
deg_df <- limma_result %>%
mutate(contrast = factor(contrast, levels = c(
"ccl_effect", "etoh_effect", "combined_effect"
))) %>%
mutate(contrast_reference = "effect")
saveRDS(deg_df, here(output_path, "limma_result.rds"))Volcano plots
Volcano plots visualizing the effect of CCl4 on gene expression.
df <- readRDS(here(output_path, "limma_result.rds"))
df %>%
filter(contrast_reference == "effect") %>%
plot_volcano() +
my_theme(grid = "y", fsize = fz)
#> filter: no rows removed
#> rename: renamed one variable (p)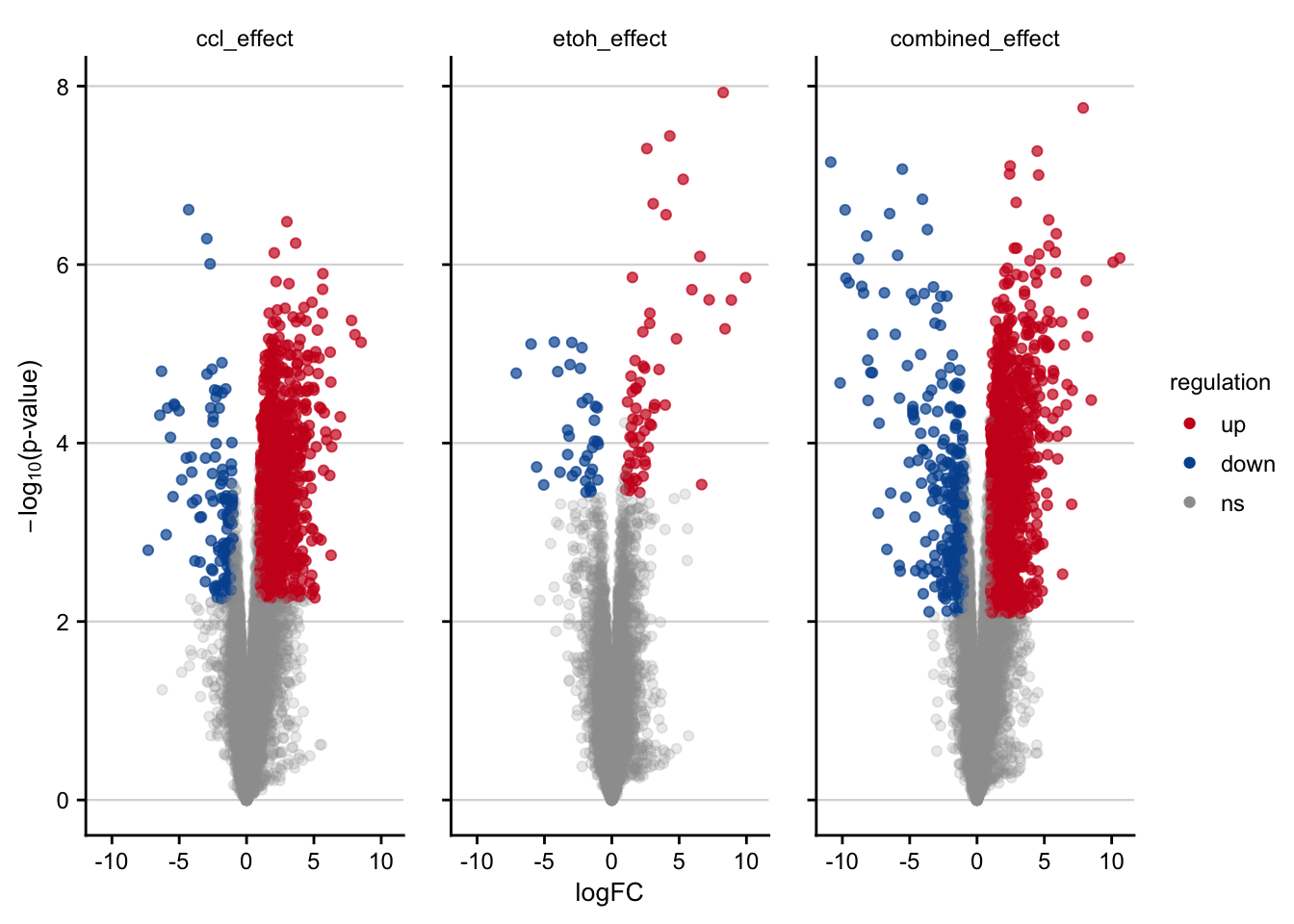
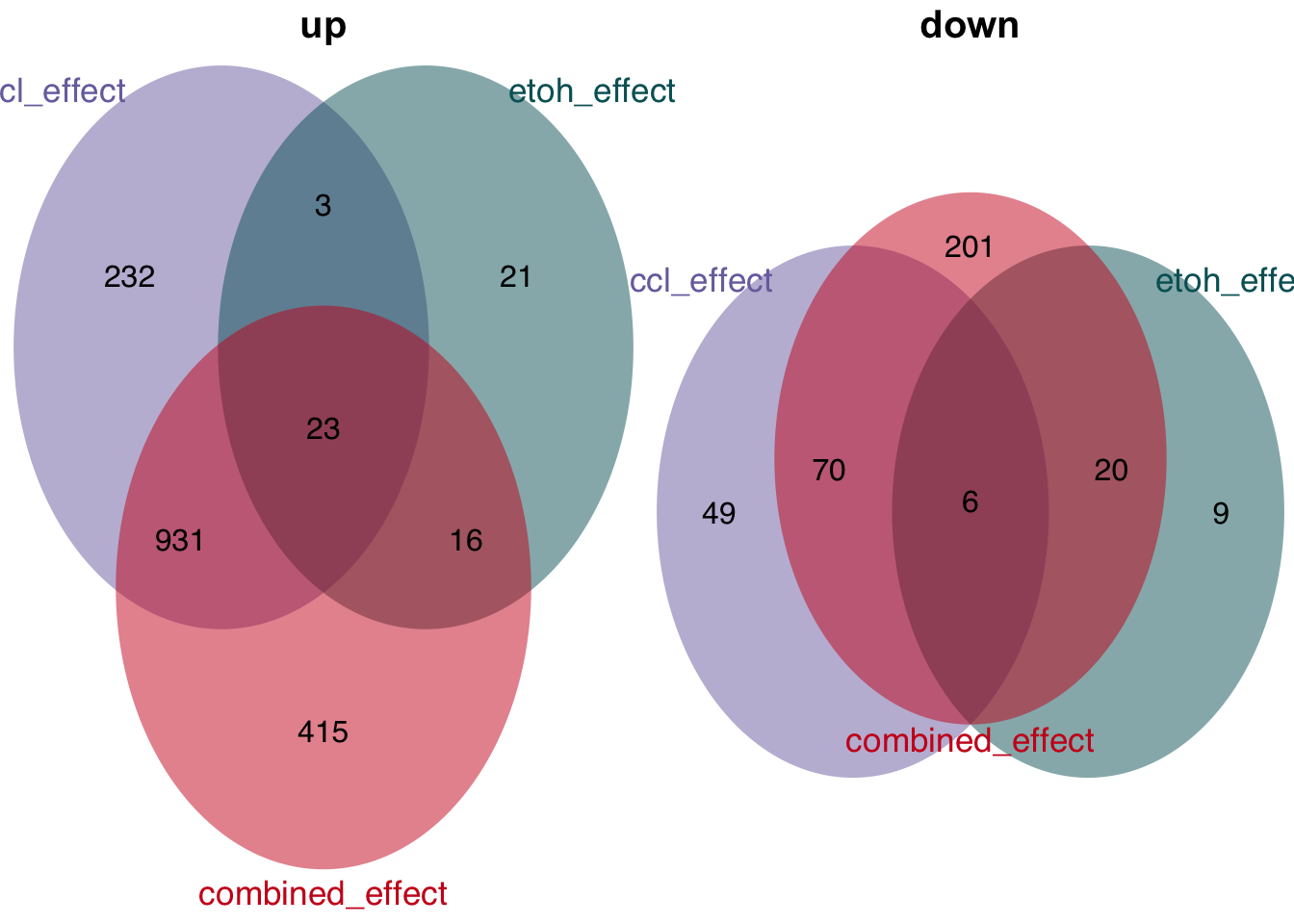
Overlap of genes
df <- readRDS(here(output_path, "limma_result.rds"))
t <- df %>%
filter(contrast_reference == "effect") %>%
rename(class = contrast) %>%
select(-contrast_reference) %>%
group_split(class)
#> filter: no rows removed
#> rename: renamed one variable (class)
#> select: dropped one variable (contrast_reference)
plot_venn_diagram(t)
#> distinct: removed 14,178 rows (>99%), one row remaining
#> distinct: removed 14,178 rows (>99%), one row remaining
#> distinct: removed 14,178 rows (>99%), one row remaining
#> count: now 3 rows and 2 columns, ungrouped
#> count: now 3 rows and 2 columns, ungrouped
#> count: now 3 rows and 2 columns, ungrouped
#> filter: removed 2 rows (67%), one row remaining
#> filter: removed 2 rows (67%), one row remaining
#> filter: removed 2 rows (67%), one row remaining
#> filter: removed 12,990 rows (92%), 1,189 rows remaining
#> filter: removed 14,116 rows (>99%), 63 rows remaining
#> filter: removed 14,116 rows (>99%), 63 rows remaining
#> filter: removed 12,794 rows (90%), 1,385 rows remaining
#> filter: removed 12,990 rows (92%), 1,189 rows remaining
#> filter: removed 12,794 rows (90%), 1,385 rows remaining
#> filter: removed 12,990 rows (92%), 1,189 rows remaining
#> filter: removed 14,116 rows (>99%), 63 rows remaining
#> filter: removed 12,794 rows (90%), 1,385 rows remaining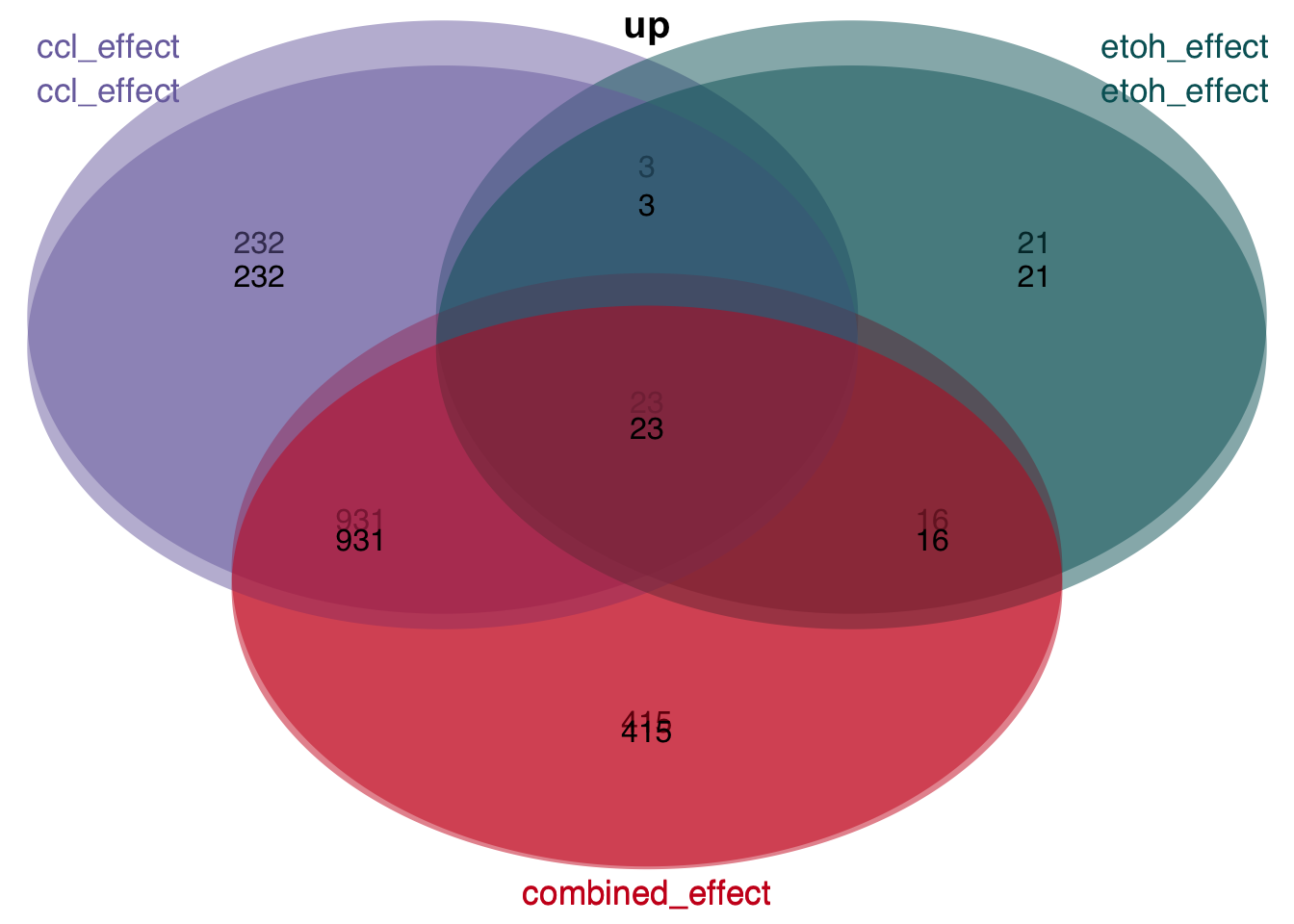
#> filter: removed 2 rows (67%), one row remaining
#> filter: removed 2 rows (67%), one row remaining
#> filter: removed 2 rows (67%), one row remaining
#> filter: removed 14,054 rows (99%), 125 rows remaining
#> filter: removed 14,144 rows (>99%), 35 rows remaining
#> filter: removed 14,144 rows (>99%), 35 rows remaining
#> filter: removed 13,882 rows (98%), 297 rows remaining
#> filter: removed 14,054 rows (99%), 125 rows remaining
#> filter: removed 13,882 rows (98%), 297 rows remaining
#> filter: removed 14,054 rows (99%), 125 rows remaining
#> filter: removed 14,144 rows (>99%), 35 rows remaining
#> filter: removed 13,882 rows (98%), 297 rows remaining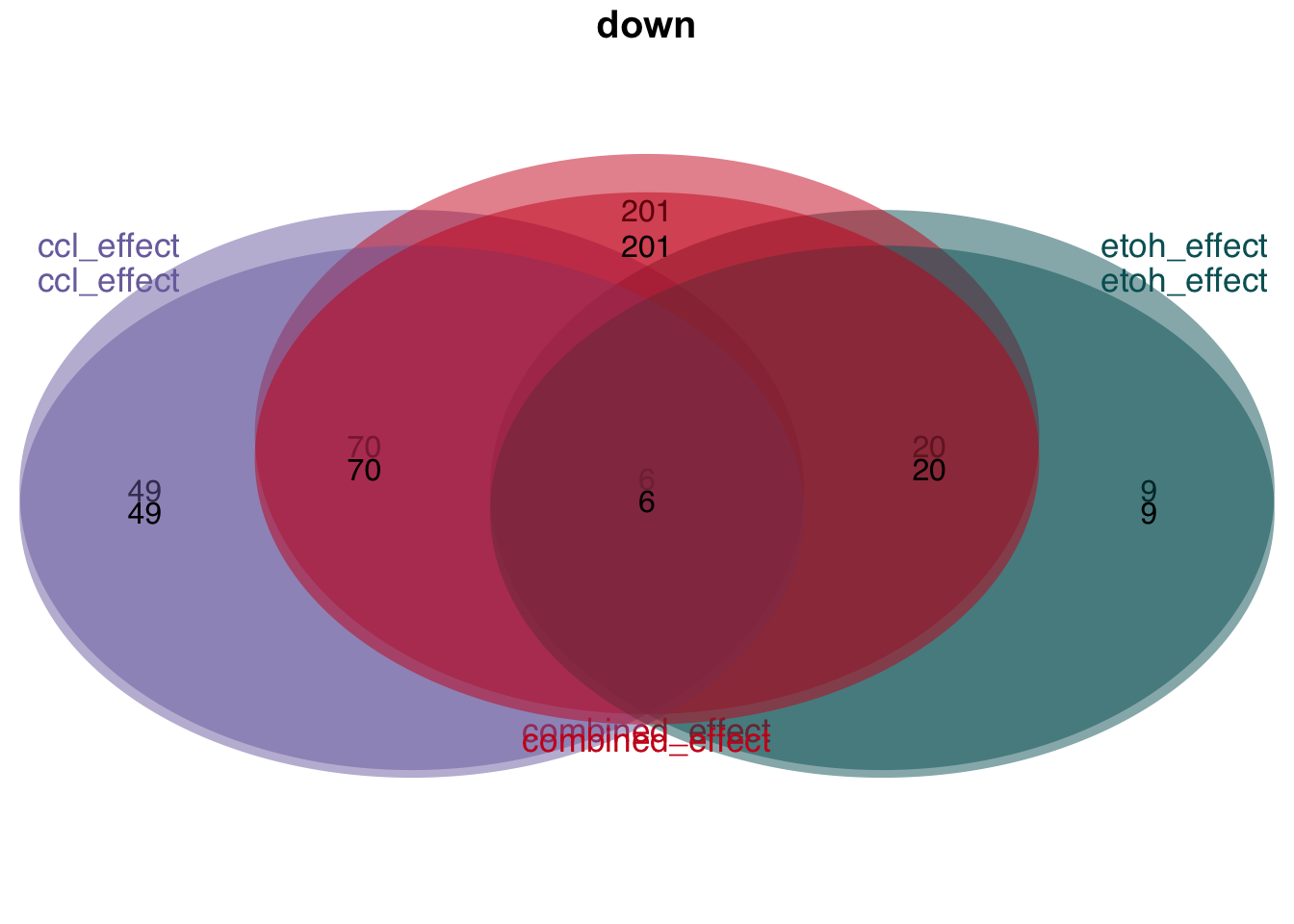
Translation to HGNC symbols
For later comparisons to human data the mouse gene symbols are mapped to their human orthologs.
df <- readRDS(here(output_path, "limma_result.rds"))
mapped_df <- df %>%
translate_gene_ids(from = "symbol_mgi", to = "symbol_hgnc") %>%
drop_na() %>%
# for duplicated genes, keep the one with the highest absolute logFC
group_by(contrast_reference, contrast, gene) %>%
slice_max(order_by = abs(logFC), n = 1, with_ties = F) %>%
ungroup()
#> select: dropped 6 variables (ensembl_mgi, ensembl_v_mgi, entrez_mgi, ensembl_hgnc, ensembl_v_hgnc, …)
#> drop_na: removed 1,905 rows (6%), 30,461 rows remaining
#> rename: renamed one variable (symbol_mgi)
#> left_join: added one column (symbol_hgnc)
#> > rows only in x 5,037
#> > rows only in y (17,348)
#> > matched rows 39,339 (includes duplicates)
#> > ========
#> > rows total 44,376
#> select: renamed one variable (gene) and dropped one variable
#> drop_na: removed 5,037 rows (11%), 39,339 rows remaining
#> slice_max (grouped): removed 1,590 rows (4%), 37,749 rows remaining
#> ungroup: no grouping variables
saveRDS(mapped_df, here(output_path, "limma_result_hs.rds"))Time spend to execute this analysis: 00:16 minutes.
sessionInfo()
#> R version 4.0.5 (2021-03-31)
#> Platform: x86_64-apple-darwin17.0 (64-bit)
#> Running under: macOS Mojave 10.14.5
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] grid stats graphics grDevices datasets utils methods
#> [8] base
#>
#> other attached packages:
#> [1] ggpubr_0.4.0 gridExtra_2.3 VennDiagram_1.6.20
#> [4] futile.logger_1.4.3 patchwork_1.1.1 lemon_0.4.5
#> [7] cowplot_1.1.0 AachenColorPalette_1.1.2 msigdf_7.1
#> [10] janitor_2.0.1 dorothea_1.0.1 progeny_1.10.0
#> [13] biobroom_1.20.0 broom_0.7.3 edgeR_3.30.3
#> [16] limma_3.44.3 here_1.0.1 tidylog_1.0.2
#> [19] forcats_0.5.0 stringr_1.4.0 dplyr_1.0.2
#> [22] purrr_0.3.4 readr_1.4.0 tidyr_1.1.2
#> [25] tibble_3.0.4 ggplot2_3.3.2 tidyverse_1.3.0
#> [28] workflowr_1.6.2
#>
#> loaded via a namespace (and not attached):
#> [1] colorspace_2.0-0 ggsignif_0.6.0 ellipsis_0.3.1
#> [4] rio_0.5.16 rprojroot_2.0.2 snakecase_0.11.0
#> [7] fs_1.5.0 rstudioapi_0.13 farver_2.0.3
#> [10] ggrepel_0.9.0 fansi_0.4.1 lubridate_1.7.9.2
#> [13] xml2_1.3.2 codetools_0.2-18 knitr_1.30
#> [16] jsonlite_1.7.2 bcellViper_1.24.0 dbplyr_2.0.0
#> [19] compiler_4.0.5 httr_1.4.2 backports_1.2.1
#> [22] assertthat_0.2.1 cli_2.2.0 later_1.1.0.1
#> [25] formatR_1.7 htmltools_0.5.0 tools_4.0.5
#> [28] gtable_0.3.0 glue_1.4.2 Rcpp_1.0.5
#> [31] carData_3.0-4 Biobase_2.48.0 cellranger_1.1.0
#> [34] vctrs_0.3.6 xfun_0.19 openxlsx_4.2.3
#> [37] rvest_0.3.6 lifecycle_0.2.0 renv_0.12.3
#> [40] rstatix_0.6.0 scales_1.1.1 clisymbols_1.2.0
#> [43] hms_0.5.3 promises_1.1.1 parallel_4.0.5
#> [46] lambda.r_1.2.4 yaml_2.2.1 curl_4.3
#> [49] stringi_1.5.3 BiocGenerics_0.34.0 zip_2.1.1
#> [52] rlang_0.4.9 pkgconfig_2.0.3 evaluate_0.14
#> [55] lattice_0.20-41 labeling_0.4.2 tidyselect_1.1.0
#> [58] plyr_1.8.6 magrittr_2.0.1 R6_2.5.0
#> [61] generics_0.1.0 DBI_1.1.0 pillar_1.4.7
#> [64] haven_2.3.1 whisker_0.4 foreign_0.8-81
#> [67] withr_2.3.0 abind_1.4-5 modelr_0.1.8
#> [70] crayon_1.3.4 car_3.0-10 futile.options_1.0.1
#> [73] rmarkdown_2.6 locfit_1.5-9.4 readxl_1.3.1
#> [76] data.table_1.13.4 git2r_0.27.1 reprex_0.3.0
#> [79] digest_0.6.27 httpuv_1.5.4 munsell_0.5.0