Knowledge-primed Neural Networks (KPNNs) for single cell data#
In this tutorial we will show how CORNETO can be used to build custom neural network architectures informed by prior knowledge. We will see how to implement a knowledge-primed neural network1. We will use the single cell data from the publication “Knowledge-primed neural networks enable biologically interpretable deep learning on single-cell sequencing data”, from Nikolaus Fortelny & Christoph Bock, where they used a single-cell RNA-seq dataset they previously generated2, which measures cellular responses to T cell receptor (TCR) stimulation in a standardized in vitro model. The dataset was chosen due to the TCR signaling pathway’s complexity and its well-characterized role in orchestrating transcriptional responses to antigen detection in T cells.
Why CORNETO?#
In the original publication, authors built a KPNN by searching on databases, building a Direct Acyclic Graph (DAG) by running shortest paths from TCR receptor to genes. However, this approach is not optimal. CORNETO, thanks to its advanced capabilities for modeling and optimization on networks, provides methods to automatically find DAG architectures in an optimal way.
In addition to this, CORNETO provides methods to build DAG NN architectures with ease using Keras +3, making KPNN implementation very flexible and interoperable with backends like Pytorch, Tensorflow and JAX.
How does it work?#
Thanks to CORNETO’s building blocks for optimization over networks, we can easily model optimization problems to find DAG architectures from a Prior Knowledge Network. After we have the backbone, we can convert it to a neural network using the utility functions included in CORNETO.
References#
Fortelny, N., & Bock, C. (2020). Knowledge-primed neural networks enable biologically interpretable deep learning on single-cell sequencing data. Genome biology, 21, 1-36.
Datlinger, P., Rendeiro, A. F., Schmidl, C., Krausgruber, T., Traxler, P., Klughammer, J., … & Bock, C. (2017). Pooled CRISPR screening with single-cell transcriptome readout. Nature methods, 14(3), 297-301.
Download and import the single cell dataset#
import os
import urllib.request
import urllib.parse
import tempfile
import pandas as pd
import scanpy as sc
import numpy as np
import corneto as cn
with urllib.request.urlopen("http://kpnn.computational-epigenetics.org/") as response:
web_input = response.geturl()
print("Effective URL:", web_input)
files = ["TCR_Edgelist.csv", "TCR_ClassLabels.csv", "TCR_Data.h5"]
temp_dir = tempfile.mkdtemp()
# Download files
file_paths = []
for file in files:
url = urllib.parse.urljoin(web_input, file)
output_path = os.path.join(temp_dir, file)
print(f"Downloading {url} to {output_path}")
try:
with urllib.request.urlopen(url) as response:
with open(output_path, 'wb') as f:
f.write(response.read())
file_paths.append(output_path)
except Exception as e:
print(f"Failed to download {url}: {e}")
print("Downloaded files:")
for path in file_paths:
print(path)
Effective URL: https://medical-epigenomics.org/papers/fortelny2019/
Downloading https://medical-epigenomics.org/papers/fortelny2019/TCR_Edgelist.csv to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpb21ulczk/TCR_Edgelist.csv
Downloading https://medical-epigenomics.org/papers/fortelny2019/TCR_ClassLabels.csv to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpb21ulczk/TCR_ClassLabels.csv
Downloading https://medical-epigenomics.org/papers/fortelny2019/TCR_Data.h5 to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpb21ulczk/TCR_Data.h5
Downloaded files:
/var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpb21ulczk/TCR_Edgelist.csv
/var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpb21ulczk/TCR_ClassLabels.csv
/var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpb21ulczk/TCR_Data.h5
# The data contains also the original network they built with shortest paths.
# We will use it to replicate the study
df_edges = pd.read_csv(file_paths[0])
df_labels = pd.read_csv(file_paths[1])
# Import the 10x data with Scanpy
adata = sc.read_10x_h5(file_paths[2])
df_labels
| barcode | TCR | |
|---|---|---|
| 0 | AAACCTGCACACATGT-1 | 0 |
| 1 | AAACCTGCACGTCTCT-1 | 0 |
| 2 | AAACCTGTCAATACCG-1 | 0 |
| 3 | AAACCTGTCGTGGTCG-1 | 0 |
| 4 | AAACGGGTCTGAGTGT-1 | 0 |
| ... | ... | ... |
| 1730 | TTTCCTCGTCATGCCG-2 | 1 |
| 1731 | TTTGCGCGTAGCCTCG-2 | 1 |
| 1732 | TTTGGTTAGATACACA-2 | 1 |
| 1733 | TTTGGTTGTATGAATG-2 | 1 |
| 1734 | TTTGGTTTCCAAGTAC-2 | 1 |
1735 rows × 2 columns
df_edges
| parent | child | |
|---|---|---|
| 0 | TCR | ZAP70 |
| 1 | ZAP70 | MAPK14 |
| 2 | MAPK14 | FOXO3 |
| 3 | MAPK14 | STAT1 |
| 4 | MAPK14 | STAT3 |
| ... | ... | ... |
| 27574 | HMGA1 | MTRNR2L9_gene |
| 27575 | MYB | C12orf50_gene |
| 27576 | MYB | TRPC5OS_gene |
| 27577 | SOX2 | TRPC5OS_gene |
| 27578 | CRTC1 | MTRNR2L9_gene |
27579 rows × 2 columns
adata.var
| gene_ids | |
|---|---|
| DDX11L1 | ENSG00000223972 |
| WASH7P | ENSG00000227232 |
| MIR6859-2 | ENSG00000278267 |
| MIR1302-10 | ENSG00000243485 |
| MIR1302-11 | ENSG00000274890 |
| ... | ... |
| Tcrlibrary_RUNX2_3_gene | Tcrlibrary_RUNX2_3_gene |
| Tcrlibrary_ZAP70_1_gene | Tcrlibrary_ZAP70_1_gene |
| Tcrlibrary_ZAP70_2_gene | Tcrlibrary_ZAP70_2_gene |
| Tcrlibrary_ZAP70_3_gene | Tcrlibrary_ZAP70_3_gene |
| Cas9_blast_gene | Cas9_blast_gene |
64370 rows × 1 columns
# We can normalize the data, however, it is better to avoid
# preprocessing the whole dataset before splitting in training and test
# to avoid data leakage.
# NOTE: Normalization can be done inside the cross-val loop
# sc.pp.normalize_total(adata, target_sum=1e6)
# Log-transform the data does not leak data as it does not estimate anything
sc.pp.log1p(adata)
adata.obs
| AAACCTGAGAAACCAT-1 |
|---|
| AAACCTGAGAAACCGC-1 |
| AAACCTGAGAAACCTA-1 |
| AAACCTGAGAAACGAG-1 |
| AAACCTGAGAAACGCC-1 |
| ... |
| TTTGTCATCTTTACAC-2 |
| TTTGTCATCTTTACGT-2 |
| TTTGTCATCTTTAGGG-2 |
| TTTGTCATCTTTAGTC-2 |
| TTTGTCATCTTTCCTC-2 |
1474560 rows × 0 columns
barcodes = adata.obs_names
barcodes
Index(['AAACCTGAGAAACCAT-1', 'AAACCTGAGAAACCGC-1', 'AAACCTGAGAAACCTA-1',
'AAACCTGAGAAACGAG-1', 'AAACCTGAGAAACGCC-1', 'AAACCTGAGAAAGTGG-1',
'AAACCTGAGAACAACT-1', 'AAACCTGAGAACAATC-1', 'AAACCTGAGAACTCGG-1',
'AAACCTGAGAACTGTA-1',
...
'TTTGTCATCTTGGGTA-2', 'TTTGTCATCTTGTACT-2', 'TTTGTCATCTTGTATC-2',
'TTTGTCATCTTGTCAT-2', 'TTTGTCATCTTGTTTG-2', 'TTTGTCATCTTTACAC-2',
'TTTGTCATCTTTACGT-2', 'TTTGTCATCTTTAGGG-2', 'TTTGTCATCTTTAGTC-2',
'TTTGTCATCTTTCCTC-2'],
dtype='object', length=1474560)
gene_names = adata.var.index
print(gene_names)
Index(['DDX11L1', 'WASH7P', 'MIR6859-2', 'MIR1302-10', 'MIR1302-11', 'FAM138A',
'OR4G4P', 'OR4G11P', 'OR4F5', 'RP11-34P13.7',
...
'Tcrlibrary_RUNX1_1_gene', 'Tcrlibrary_RUNX1_2_gene',
'Tcrlibrary_RUNX1_3_gene', 'Tcrlibrary_RUNX2_1_gene',
'Tcrlibrary_RUNX2_2_gene', 'Tcrlibrary_RUNX2_3_gene',
'Tcrlibrary_ZAP70_1_gene', 'Tcrlibrary_ZAP70_2_gene',
'Tcrlibrary_ZAP70_3_gene', 'Cas9_blast_gene'],
dtype='object', length=64370)
len(set(df_labels.barcode.tolist()))
1735
len(set(barcodes.tolist()))
1474560
matched_barcodes = sorted(set(barcodes.tolist()) & set(df_labels.barcode.tolist()))
len(matched_barcodes)
1735
#This is the InPathsY data in the original code of KPNNs
df_labels
| barcode | TCR | |
|---|---|---|
| 0 | AAACCTGCACACATGT-1 | 0 |
| 1 | AAACCTGCACGTCTCT-1 | 0 |
| 2 | AAACCTGTCAATACCG-1 | 0 |
| 3 | AAACCTGTCGTGGTCG-1 | 0 |
| 4 | AAACGGGTCTGAGTGT-1 | 0 |
| ... | ... | ... |
| 1730 | TTTCCTCGTCATGCCG-2 | 1 |
| 1731 | TTTGCGCGTAGCCTCG-2 | 1 |
| 1732 | TTTGGTTAGATACACA-2 | 1 |
| 1733 | TTTGGTTGTATGAATG-2 | 1 |
| 1734 | TTTGGTTTCCAAGTAC-2 | 1 |
1735 rows × 2 columns
Import PKN with CORNETO#
import corneto as cn
cn.info()
|
|
|
outputs_pkn = list(set(df_edges.parent.tolist()) - set(df_edges.child.tolist()))
inputs_pkn = set(df_edges.child.tolist()) - set(df_edges.parent.tolist())
input_pkn_genes = list(set(g.split("_")[0] for g in inputs_pkn))
len(inputs_pkn), len(outputs_pkn)
(13121, 1)
tuples = [(r.child, 1, r.parent) for _, r in df_edges.iterrows()]
G = cn.Graph.from_sif_tuples(tuples)
G = G.prune(inputs_pkn, outputs_pkn)
# Size of the original PKN provided by the authors
G.shape
(13439, 27579)
Select the single cell data for training#
adata_matched = adata[adata.obs_names.isin(matched_barcodes), adata.var_names.isin(input_pkn_genes)]
adata_matched.shape
(1735, 14229)
non_zero_genes = set(adata_matched.to_df().columns[adata_matched.to_df().sum(axis=0) >= 1e-6].values)
len(non_zero_genes)
12459
len(non_zero_genes.intersection(adata_matched.var_names))
12459
adata_matched = adata_matched[:, adata_matched.var_names.isin(non_zero_genes)]
# Many duplicates still 0 counts
adata_matched = adata_matched[:, adata_matched.to_df().sum(axis=0) != 0]
adata_matched.shape
(1735, 12487)
df_expr = adata_matched.to_df()
df_expr = df_expr.groupby(df_expr.columns, axis=1).max()
df_expr
| A1BG | A2ML1 | AAAS | AACS | AADAT | AAED1 | AAGAB | AAK1 | AAMDC | AAMP | ... | ZSWIM8 | ZUFSP | ZW10 | ZWILCH | ZXDC | ZYG11A | ZYG11B | ZYX | ZZEF1 | ZZZ3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAACCTGCACACATGT-1 | 0.0 | 0.0 | 0.000000 | 0.693147 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 1.098612 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 |
| AAACCTGCACGTCTCT-1 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.0 | 0.693147 | 0.000000 | 0.000000 | 0.0 | 0.693147 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 |
| AAACCTGTCAATACCG-1 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.693147 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 |
| AAACCTGTCGTGGTCG-1 | 0.0 | 0.0 | 1.098612 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 1.098612 | ... | 0.000000 | 0.693147 | 0.000000 | 0.693147 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 |
| AAACGGGTCTGAGTGT-1 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.0 | 0.693147 | 0.000000 | 0.000000 | 0.0 | 0.000000 | ... | 0.693147 | 0.000000 | 0.693147 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| TTTCCTCGTCATGCCG-2 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.693147 | 0.0 | 0.693147 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 |
| TTTGCGCGTAGCCTCG-2 | 0.0 | 0.0 | 1.098612 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 1.386294 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 |
| TTTGGTTAGATACACA-2 | 0.0 | 0.0 | 1.098612 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 1.098612 | 0.0 | 0.693147 | ... | 0.693147 | 0.000000 | 0.693147 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.693147 | 0.693147 | 0.000000 |
| TTTGGTTGTATGAATG-2 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.0 | 0.693147 | 1.098612 | 0.000000 | 0.0 | 0.693147 | ... | 0.000000 | 0.693147 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.693147 |
| TTTGGTTTCCAAGTAC-2 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.693147 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 1.098612 | 0.000000 | 0.000000 |
1735 rows × 12459 columns
Building and training the KPNN#
Now we will use the provided PKN by the authors and the utility functions in CORNETO to build a KPNN similar to the one used in the original manuscript
import os
os.environ["KERAS_BACKEND"] = "jax"
import keras
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score, roc_auc_score
from keras.optimizers import Adam
from keras.callbacks import EarlyStopping
import numpy as np
# Use the data from the experiment
X = df_expr.values
y = df_labels.set_index("barcode").loc[df_expr.index, "TCR"].values
X.shape, y.shape
((1735, 12459), (1735,))
# We can prefilter on top N genes to make this faster
top_n = None
# From the given PKN
outputs_pkn = list(set(df_edges.parent.tolist()) - set(df_edges.child.tolist()))
inputs_pkn = set(df_edges.child.tolist()) - set(df_edges.parent.tolist())
input_pkn_genes = list(set(g.split("_")[0] for g in inputs_pkn))
if top_n is not None and top_n > 0:
input_pkn_genes = list(set(input_pkn_genes).intersection(df_expr.var(axis=0).sort_values(ascending=False).head(top_n).index))
inputs_pkn = list(g + "_gene" for g in input_pkn_genes)
len(inputs_pkn), len(outputs_pkn)
(13121, 1)
input_nn_genes = list(set(input_pkn_genes).intersection(df_expr.columns))
input_nn = [g + "_gene" for g in input_nn_genes]
len(input_nn)
12459
# Build corneto graph
tuples = [(r.child, 1, r.parent) for _, r in df_edges.iterrows()]
G = cn.Graph.from_sif_tuples(tuples)
G = G.prune(input_nn, outputs_pkn)
G.shape
(12767, 25928)
len(input_nn), len(input_nn_genes)
(12459, 12459)
len(set(input_nn).intersection(G.V))
12459
X = df_expr.loc[:, input_nn_genes].values
y = df_labels.set_index("barcode").loc[df_expr.index, "TCR"].values
X.shape, y.shape
((1735, 12459), (1735,))
from corneto._ml import build_dagnn
def stratified_kfold(
G,
inputs,
outputs,
n_splits=5,
shuffle=True,
random_state=42,
lr=0.001,
patience=10,
file_weights="weights",
dagnn_config=dict(
batch_norm_input=True,
batch_norm_center=False,
batch_norm_scale=False,
bias_reg_l1=1e-3,
bias_reg_l2=1e-2,
dropout=0.20,
default_hidden_activation="sigmoid",
default_output_activation="sigmoid",
verbose=False
)
):
kfold = StratifiedKFold(n_splits=n_splits, shuffle=shuffle, random_state=random_state)
models = []
metrics = {m: [] for m in ["accuracy", "precision", "recall", "f1", "roc_auc"]}
for i, (train_idx, val_idx) in enumerate(kfold.split(X, y)):
X_train, X_val = X[train_idx], X[val_idx]
y_train, y_val = y[train_idx], y[val_idx]
print("Building DAG NN model with CORNETO using Keras with JAX...")
print(f" > N. inputs: {len(input_nn)}")
print(f" > N. outputs: {len(outputs_pkn)}")
model = build_dagnn(
G,
input_nn,
outputs_pkn,
**dagnn_config
)
print(f" > N. parameters: {model.count_params()}")
# Train the model with Adam
opt=keras.optimizers.Adam(learning_rate=lr)
early_stopping = EarlyStopping(monitor='val_loss', patience=patience, restore_best_weights=True)
print("Compiling...")
model.compile(
optimizer=opt,
loss='binary_crossentropy',
metrics=['accuracy']
)
print("Fitting...")
model.fit(X_train, y_train,
validation_data=(X_val, y_val),
epochs=200,
batch_size=64,
verbose=0,
callbacks=[early_stopping])
if file_weights is not None:
filename = f"{file_weights}_{i}.keras"
model.save(filename)
print(f"Weights saved to {filename}")
# Predictions and metrics calculation
y_pred_proba = model.predict(X_val).flatten()
y_pred = (y_pred_proba > 0.5).astype(int)
acc = accuracy_score(y_val, y_pred)
precision = precision_score(y_val, y_pred)
recall = recall_score(y_val, y_pred)
f1 = f1_score(y_val, y_pred)
roc_auc = roc_auc_score(y_val, y_pred_proba)
metrics["accuracy"].append(acc)
metrics["precision"].append(precision)
metrics["recall"].append(recall)
metrics["f1"].append(f1)
metrics["roc_auc"].append(roc_auc)
print(f" > Fold {i} validation ROC-AUC={roc_auc:.3f}")
models.append(model)
return models, metrics
temp_weights = tempfile.mkdtemp()
models, metrics = stratified_kfold(G, input_nn, outputs_pkn, file_weights=os.path.join(temp_weights, "weights"))
print("Validation metrics:")
for k, v in metrics.items():
print(f" - {k}: {np.mean(v):.3f}")
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 26236
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/weights_0.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 4s 208ms/step
> Fold 0 validation ROC-AUC=0.991
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 26236
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/weights_1.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 4s 180ms/step
> Fold 1 validation ROC-AUC=0.983
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 26236
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/weights_2.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 4s 179ms/step
> Fold 2 validation ROC-AUC=0.986
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 26236
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/weights_3.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 4s 180ms/step
> Fold 3 validation ROC-AUC=0.993
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 26236
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/weights_4.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 4s 181ms/step
> Fold 4 validation ROC-AUC=0.994
Validation metrics:
- accuracy: 0.957
- precision: 0.961
- recall: 0.951
- f1: 0.956
- roc_auc: 0.989
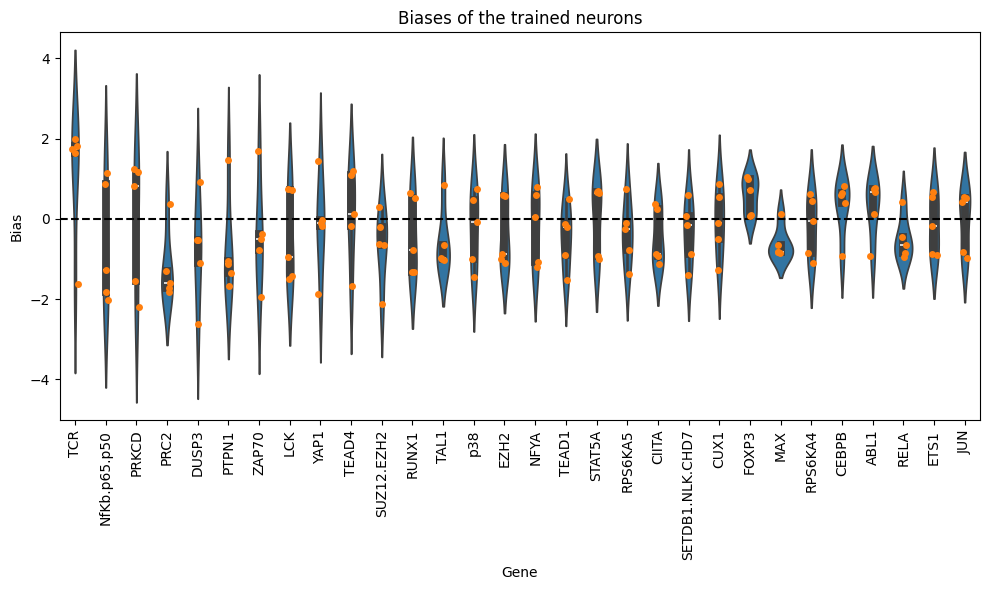
Now, we will analyze the learned biases for each of the nodes in the graph. Note that authors in the KPNN paper explain a way to extract weights for the nodes, based on the learned interactions and accounting for biases in the structure of the NN. Here we just show the learned biases of the nodes of the NN across 5 folds. Please be careful interpreting these weights.
# We collect the weights obtained in each fold
def load_biases(file="weights", folds=5):
biases = []
mean_inputs = []
for i in range(5):
model = keras.models.load_model(f"{file}_{i}.keras")
for layer in model.layers:
weights = layer.get_weights()
if weights:
biases.append((i, layer.name, weights[1][0]))
mean_inputs.append((i, layer.name, weights[0].mean()))
df_biases = pd.DataFrame(biases, columns=["fold", "gene", "bias"])
df_biases["abs_bias"] = df_biases.bias.abs()
df_biases["pow2_bias"] = df_biases.bias.pow(2)
df_biases = df_biases.set_index(["fold","gene"])
return df_biases
df_biases = load_biases(file=os.path.join(temp_weights, "weights"), folds=5)
df_biases.sort_values(by="abs_bias", ascending=False).head(10)
| bias | abs_bias | pow2_bias | ||
|---|---|---|---|---|
| fold | gene | |||
| 1 | DUSP3 | -2.632734 | 2.632734 | 6.931287 |
| 2 | PRKCD | -2.197672 | 2.197672 | 4.829763 |
| SUZ12.EZH2 | -2.126150 | 2.126150 | 4.520514 | |
| NfKb.p65.p50 | -2.010682 | 2.010682 | 4.042840 | |
| TCR | 1.988334 | 1.988334 | 3.953472 | |
| 4 | ZAP70 | -1.952261 | 1.952261 | 3.811324 |
| 3 | YAP1 | -1.872004 | 1.872004 | 3.504399 |
| 4 | NfKb.p65.p50 | -1.834747 | 1.834747 | 3.366298 |
| 0 | PRC2 | -1.826627 | 1.826627 | 3.336568 |
| 4 | TCR | 1.816516 | 1.816516 | 3.299730 |
df_biases_full = df_biases.copy().reset_index()
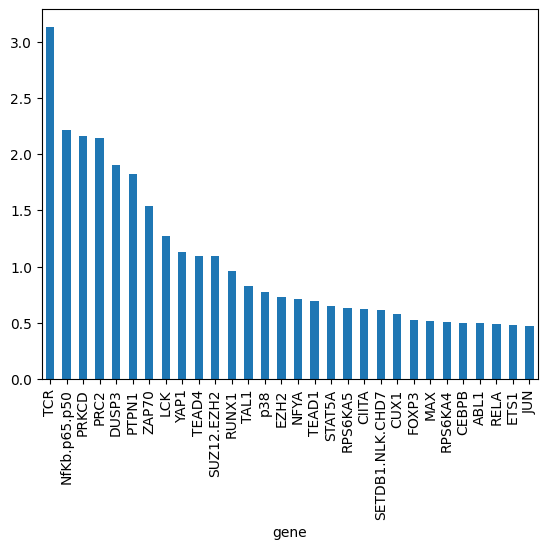
gene_biases_score = df_biases_full.groupby("gene")["pow2_bias"].mean().sort_values(ascending=False)
gene_biases_score
gene
TCR 3.129825
NfKb.p65.p50 2.217130
PRKCD 2.162571
PRC2 2.140556
DUSP3 1.901882
...
MLL2.complex 0.000641
HSF2 0.000268
GATA3 0.000145
SRY 0.000138
REST 0.000032
Name: pow2_bias, Length: 308, dtype: float32
gene_biases_score.head(30).plot.bar()
<Axes: xlabel='gene'>
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# Get the top genes sorted by mean bias
top_genes = gene_biases_score.head(30).index
# Filter the DataFrame to include only rows with these top genes
filtered_df = df_biases_full[df_biases_full['gene'].isin(top_genes)]
plt.figure(figsize=(10, 6))
sns.violinplot(data=filtered_df, x="gene", y="bias", order=top_genes, dodge=True)
sns.stripplot(data=filtered_df, x="gene", y="bias", order=top_genes, dodge=True)
plt.xticks(rotation=90)
plt.title('Biases of the trained neurons')
plt.xlabel('Gene')
plt.ylabel('Bias')
plt.axhline(0, linestyle="--", color="k")
plt.tight_layout()
Use CORNETO for NN pruning#
Now we will show how CORNETO can be used to extract a smaller, yet complete DAG from the original PKN provided by the authors. We will add input edges to each input node and an output edge through TCR to indicate which nodes are the inputs and which one the output. We will use then Acyclic Flow to find the smallest DAG comprising these nodes
G_dag = G.copy()
new_edges = []
for g in input_nn:
new_edges.append(G_dag.add_edge((), g))
new_edges.append(G_dag.add_edge("TCR", ()))
print(G_dag.shape)
# Find small DAG. We use Acyclic Flow to find over the space of DAGs
P = cn.opt.AcyclicFlow(G_dag)
# We enforce that the input genes and the output gene are part of the solution
P += P.expr.with_flow[new_edges] == 1
# Minimize the number of active edges
P.add_objectives(sum(P.expr.with_flow), weights=1)
P.solve(solver="SCIP", verbosity=1, max_seconds=300);
(12767, 38388)
===============================================================================
CVXPY
v1.6.0
===============================================================================
(CVXPY) Dec 23 01:19:36 PM: Your problem has 127931 variables, 332945 constraints, and 0 parameters.
(CVXPY) Dec 23 01:19:36 PM: It is compliant with the following grammars: DCP, DQCP
(CVXPY) Dec 23 01:19:36 PM: (If you need to solve this problem multiple times, but with different data, consider using parameters.)
(CVXPY) Dec 23 01:19:36 PM: CVXPY will first compile your problem; then, it will invoke a numerical solver to obtain a solution.
(CVXPY) Dec 23 01:19:36 PM: Your problem is compiled with the CPP canonicalization backend.
-------------------------------------------------------------------------------
Compilation
-------------------------------------------------------------------------------
(CVXPY) Dec 23 01:19:37 PM: Compiling problem (target solver=SCIP).
(CVXPY) Dec 23 01:19:37 PM: Reduction chain: Dcp2Cone -> CvxAttr2Constr -> ConeMatrixStuffing -> SCIP
(CVXPY) Dec 23 01:19:37 PM: Applying reduction Dcp2Cone
(CVXPY) Dec 23 01:19:37 PM: Applying reduction CvxAttr2Constr
(CVXPY) Dec 23 01:19:37 PM: Applying reduction ConeMatrixStuffing
(CVXPY) Dec 23 01:20:56 PM: Applying reduction SCIP
(CVXPY) Dec 23 01:20:56 PM: Finished problem compilation (took 7.962e+01 seconds).
-------------------------------------------------------------------------------
Numerical solver
-------------------------------------------------------------------------------
(CVXPY) Dec 23 01:20:56 PM: Invoking solver SCIP to obtain a solution.
presolving:
(round 1, fast) 72218 del vars, 263764 del conss, 0 add conss, 158463 chg bounds, 17010 chg sides, 17010 chg coeffs, 0 upgd conss, 0 impls, 0 clqs
(round 2, fast) 83111 del vars, 288686 del conss, 0 add conss, 158636 chg bounds, 17982 chg sides, 17982 chg coeffs, 0 upgd conss, 0 impls, 0 clqs
(round 3, fast) 83299 del vars, 288858 del conss, 0 add conss, 158667 chg bounds, 18157 chg sides, 18150 chg coeffs, 0 upgd conss, 0 impls, 0 clqs
(round 4, fast) 83323 del vars, 288959 del conss, 0 add conss, 158670 chg bounds, 18308 chg sides, 18242 chg coeffs, 0 upgd conss, 0 impls, 0 clqs
(round 5, fast) 87954 del vars, 288998 del conss, 0 add conss, 158670 chg bounds, 18310 chg sides, 18242 chg coeffs, 0 upgd conss, 0 impls, 0 clqs
(round 6, fast) 87954 del vars, 288998 del conss, 0 add conss, 169675 chg bounds, 18310 chg sides, 18242 chg coeffs, 0 upgd conss, 0 impls, 0 clqs
(round 7, fast) 87976 del vars, 289373 del conss, 0 add conss, 169675 chg bounds, 19713 chg sides, 29203 chg coeffs, 0 upgd conss, 0 impls, 0 clqs
(round 8, fast) 87993 del vars, 289470 del conss, 0 add conss, 169675 chg bounds, 19713 chg sides, 29203 chg coeffs, 0 upgd conss, 0 impls, 0 clqs
(round 9, exhaustive) 87994 del vars, 300376 del conss, 0 add conss, 169708 chg bounds, 19713 chg sides, 29203 chg coeffs, 0 upgd conss, 0 impls, 0 clqs
(round 10, fast) 98924 del vars, 311359 del conss, 0 add conss, 169708 chg bounds, 19722 chg sides, 29210 chg coeffs, 0 upgd conss, 4004 impls, 0 clqs
(round 11, fast) 99966 del vars, 312534 del conss, 0 add conss, 169708 chg bounds, 19722 chg sides, 29210 chg coeffs, 0 upgd conss, 4004 impls, 0 clqs
(round 12, fast) 99978 del vars, 312581 del conss, 0 add conss, 169709 chg bounds, 19722 chg sides, 29210 chg coeffs, 0 upgd conss, 4004 impls, 0 clqs
(round 13, exhaustive) 99980 del vars, 312589 del conss, 0 add conss, 169715 chg bounds, 19724 chg sides, 29212 chg coeffs, 9689 upgd conss, 4004 impls, 0 clqs
(round 14, fast) 99988 del vars, 312611 del conss, 0 add conss, 169715 chg bounds, 19724 chg sides, 29212 chg coeffs, 9689 upgd conss, 14702 impls, 925 clqs
(round 15, exhaustive) 99999 del vars, 312621 del conss, 0 add conss, 169718 chg bounds, 19726 chg sides, 29214 chg coeffs, 17411 upgd conss, 14704 impls, 908 clqs
(round 16, medium) 100057 del vars, 312642 del conss, 0 add conss, 169718 chg bounds, 19726 chg sides, 29214 chg coeffs, 17411 upgd conss, 22425 impls, 1333 clqs
(round 17, medium) 100059 del vars, 312756 del conss, 0 add conss, 169718 chg bounds, 19726 chg sides, 29214 chg coeffs, 17411 upgd conss, 22425 impls, 1333 clqs
(round 18, exhaustive) 100754 del vars, 312760 del conss, 0 add conss, 169720 chg bounds, 19726 chg sides, 29214 chg coeffs, 17413 upgd conss, 22425 impls, 1333 clqs
(round 19, fast) 100756 del vars, 312828 del conss, 0 add conss, 169720 chg bounds, 19726 chg sides, 29219 chg coeffs, 17413 upgd conss, 22427 impls, 1337 clqs
(3.0s) probing: 1000/17563 (5.7%) - 0 fixings, 1 aggregations, 370 implications, 2 bound changes
(3.0s) probing: 1003/17563 (5.7%) - 0 fixings, 1 aggregations, 370 implications, 2 bound changes
(3.0s) probing aborted: 1000/1000 successive useless probings
(3.0s) symmetry computation started: requiring (bin +, int +, cont +), (fixed: bin -, int -, cont -)
(26.3s) symmetry computation finished: 1500 generators found (max: 1500, log10 of symmetry group size: 1750.0) (symcode time: 23.21)
dynamic symmetry handling statistics:
orbitopal reduction: 5 components: 3x3, 3x3, 3x3, 3x3, 4x3
orbital reduction: 11 components of sizes 3, 5, 9, 4, 4, 4, 18, 17, 8, 6, 12
lexicographic reduction: 105 permutations with support sizes 8, 6, 8, 6, 6, 8, 6, 6, 6, 8, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 8, 8, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 8, 8, 8, 8, 8, 8, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6
handled 45 out of 45 symmetry components
(round 20, exhaustive) 100757 del vars, 312828 del conss, 1385 add conss, 169722 chg bounds, 19726 chg sides, 29219 chg coeffs, 17417 upgd conss, 22520 impls, 1612 clqs
(round 21, exhaustive) 101738 del vars, 312830 del conss, 1385 add conss, 169722 chg bounds, 19726 chg sides, 29219 chg coeffs, 17417 upgd conss, 22528 impls, 1616 clqs
(round 22, fast) 101738 del vars, 313811 del conss, 1385 add conss, 169722 chg bounds, 19726 chg sides, 29219 chg coeffs, 17417 upgd conss, 22528 impls, 1616 clqs
(26.5s) probing: 1103/17563 (6.3%) - 0 fixings, 1 aggregations, 563 implications, 2 bound changes
(26.5s) probing aborted: 1000/1000 successive useless probings
presolving (23 rounds: 23 fast, 9 medium, 7 exhaustive):
101738 deleted vars, 313811 deleted constraints, 1385 added constraints, 169722 tightened bounds, 0 added holes, 19726 changed sides, 29219 changed coefficients
22621 implications, 1716 cliques
presolved problem has 26193 variables (17562 bin, 0 int, 0 impl, 8631 cont) and 20519 constraints
16888 constraints of type <varbound>
3323 constraints of type <linear>
308 constraints of type <logicor>
transformed objective value is always integral (scale: 1)
Presolving Time: 26.29
time | node | left |LP iter|LP it/n|mem/heur|mdpt |vars |cons |rows |cuts |sepa|confs|strbr| dualbound | primalbound | gap | compl.
p26.7s| 1 | 0 | 115 | - | locks| 0 | 26k| 20k| 20k| 0 | 0 | 0 | 0 | 2.066500e+04 | 2.744200e+04 | 32.79%| unknown
i26.8s| 1 | 0 | 115 | - | oneopt| 0 | 26k| 20k| 20k| 0 | 0 | 24 | 0 | 2.066500e+04 | 2.707300e+04 | 31.01%| unknown
27.6s| 1 | 0 | 11865 | - | 661M | 0 | 26k| 20k| 20k| 0 | 0 | 24 | 0 | 2.097314e+04 | 2.707300e+04 | 29.08%| unknown
28.4s| 1 | 0 | 16422 | - | 671M | 0 | 26k| 20k| 21k|1624 | 1 | 24 | 0 | 2.258309e+04 | 2.707300e+04 | 19.88%| unknown
29.4s| 1 | 0 | 21208 | - | 674M | 0 | 26k| 20k| 21k|1857 | 2 | 24 | 0 | 2.279908e+04 | 2.707300e+04 | 18.75%| unknown
29.7s| 1 | 0 | 23243 | - | 676M | 0 | 26k| 20k| 22k|2002 | 3 | 24 | 0 | 2.292005e+04 | 2.707300e+04 | 18.12%| unknown
r29.8s| 1 | 0 | 23243 | - |shifting| 0 | 26k| 20k| 22k|2002 | 3 | 24 | 0 | 2.292005e+04 | 2.515100e+04 | 9.73%| unknown
30.2s| 1 | 0 | 25780 | - | 680M | 0 | 26k| 20k| 22k|2120 | 4 | 24 | 0 | 2.302305e+04 | 2.515100e+04 | 9.24%| unknown
i32.7s| 1 | 0 | 37528 | - | oneopt| 0 | 26k| 20k| 22k|2120 | 4 | 24 | 0 | 2.302305e+04 | 2.514600e+04 | 9.22%| unknown
(node 1) unresolved numerical troubles in LP 10 -- using pseudo solution instead (loop 1)
35.8s| 1 | 2 | 49334 | - | 684M | 0 | 26k| 20k| 22k|2120 | 4 | 24 | 0 | 2.302305e+04 | 2.514600e+04 | 9.22%| unknown
d72.5s| 68 | 69 | 98165 | 730.5 |veclendi| 11 | 26k| 20k| 22k| 0 | 1 | 24 |1210 | 2.302805e+04 | 2.511900e+04 | 9.08%| unknown
o82.1s| 92 | 93 |122341 | 803.5 |rootsold| 13 | 26k| 20k| 22k|2233 | 1 | 24 |1568 | 2.302805e+04 | 2.509600e+04 | 8.98%| unknown
84.6s| 100 | 101 |127795 | 793.7 | 734M | 13 | 26k| 20k| 22k|2251 | 1 | 24 |1659 | 2.302805e+04 | 2.509600e+04 | 8.98%| unknown
d92.0s| 125 | 126 |148749 | 802.7 |guideddi| 15 | 26k| 20k| 22k| 0 | 1 | 24 |1833 | 2.303005e+04 | 2.508900e+04 | 8.94%| unknown
r 114s| 190 | 191 |204186 | 819.9 |ziroundi| 17 | 26k| 20k| 22k|2380 | 1 | 24 |2696 | 2.303205e+04 | 2.508700e+04 | 8.92%| unknown
time | node | left |LP iter|LP it/n|mem/heur|mdpt |vars |cons |rows |cuts |sepa|confs|strbr| dualbound | primalbound | gap | compl.
118s| 200 | 201 |209863 | 807.3 | 775M | 17 | 26k| 20k| 22k|2406 | 1 | 24 |2831 | 2.303304e+04 | 2.508700e+04 | 8.92%| unknown
r 139s| 262 | 263 |255895 | 791.9 |ziroundi| 20 | 26k| 20k| 22k|2504 | 1 | 28 |3608 | 2.303304e+04 | 2.508500e+04 | 8.91%| unknown
r 139s| 263 | 264 |255962 | 789.1 |ziroundi| 21 | 26k| 20k| 22k|2504 | 1 | 28 |3628 | 2.303304e+04 | 2.508400e+04 | 8.90%| unknown
r 148s| 297 | 298 |269173 | 743.1 |ziroundi| 21 | 26k| 20k| 22k|2538 | 1 | 29 |4061 | 2.303404e+04 | 2.508400e+04 | 8.90%| unknown
150s| 300 | 301 |274671 | 754.0 | 818M | 21 | 26k| 20k| 22k|2538 | 1 | 30 |4103 | 2.303404e+04 | 2.508400e+04 | 8.90%| unknown
r 154s| 317 | 318 |279506 | 728.8 |ziroundi| 21 | 26k| 20k| 22k|2555 | 1 | 30 |4323 | 2.303404e+04 | 2.508400e+04 | 8.90%| unknown
r 155s| 322 | 323 |279561 | 717.6 |ziroundi| 21 | 26k| 20k| 22k|2555 | 1 | 30 |4365 | 2.303404e+04 | 2.508300e+04 | 8.90%| unknown
r 171s| 390 | 391 |313739 | 680.0 |ziroundi| 21 | 26k| 20k| 22k|2602 | 1 | 35 |5079 | 2.303404e+04 | 2.508300e+04 | 8.90%| unknown
174s| 400 | 401 |320440 | 679.8 | 881M | 21 | 26k| 20k| 22k|2603 | 1 | 55 |5167 | 2.303404e+04 | 2.508300e+04 | 8.90%| unknown
r 177s| 418 | 419 |326124 | 664.0 |ziroundi| 21 | 26k| 20k| 22k|2603 | 1 | 56 |5295 | 2.303404e+04 | 2.508300e+04 | 8.90%| unknown
202s| 500 | 501 |405291 | 713.6 | 909M | 21 | 26k| 20k| 22k|2734 | 1 | 65 |5922 | 2.303504e+04 | 2.508300e+04 | 8.89%| unknown
226s| 600 | 601 |476672 | 713.6 | 922M | 23 | 26k| 20k| 22k|2845 | 2 | 73 |6493 | 2.303504e+04 | 2.508300e+04 | 8.89%| unknown
L 230s| 618 | 619 |481926 | 701.3 | rins| 23 | 26k| 20k| 22k|2853 | 1 | 73 |6520 | 2.303504e+04 | 2.508200e+04 | 8.89%| unknown
244s| 700 | 701 |519687 | 673.1 | 932M | 23 | 26k| 20k| 22k|2943 | 1 | 77 |6751 | 2.303505e+04 | 2.508200e+04 | 8.89%| unknown
258s| 800 | 801 |554187 | 632.0 | 936M | 25 | 26k| 20k| 22k|3050 | 2 | 77 |7000 | 2.303602e+04 | 2.508200e+04 | 8.88%| unknown
time | node | left |LP iter|LP it/n|mem/heur|mdpt |vars |cons |rows |cuts |sepa|confs|strbr| dualbound | primalbound | gap | compl.
r 271s| 860 | 861 |603796 | 645.6 |ziroundi| 25 | 26k| 20k| 22k|3093 | 1 | 78 |7085 | 2.303602e+04 | 2.508200e+04*| 8.88%| unknown
r 272s| 874 | 875 |605526 | 637.2 |ziroundi| 25 | 26k| 20k| 22k|3093 | 1 | 78 |7096 | 2.303604e+04 | 2.508000e+04 | 8.87%| unknown
r 272s| 887 | 888 |609332 | 632.2 |ziroundi| 25 | 26k| 20k| 22k|3102 | 1 | 78 |7102 | 2.303604e+04 | 2.508000e+04 | 8.87%| unknown
r 273s| 888 | 889 |611391 | 633.8 |ziroundi| 25 | 26k| 20k| 22k|3102 | 1 | 78 |7102 | 2.303604e+04 | 2.507900e+04 | 8.87%| unknown
273s| 900 | 901 |613629 | 627.8 | 965M | 25 | 26k| 20k| 22k|3102 | 1 | 78 |7119 | 2.303604e+04 | 2.507900e+04 | 8.87%| unknown
290s| 1000 | 1001 |659345 | 610.7 | 971M | 25 | 26k| 20k| 22k|3208 | 1 | 81 |7495 | 2.303604e+04 | 2.507900e+04 | 8.87%| unknown
r 298s| 1044 | 1045 |672493 | 597.6 |ziroundi| 27 | 26k| 20k| 22k|3251 | 1 | 81 |7598 | 2.303604e+04 | 2.507800e+04 | 8.86%| unknown
Restart triggered after 50 consecutive estimations that the remaining tree will be large
(run 1, node 1049) performing user restart
(restart) converted 1743 cuts from the global cut pool into linear constraints
presolving:
(round 1, exhaustive) 0 del vars, 10 del conss, 0 add conss, 0 chg bounds, 0 chg sides, 0 chg coeffs, 1721 upgd conss, 22621 impls, 1903 clqs
(round 2, medium) 0 del vars, 20 del conss, 10 add conss, 2 chg bounds, 0 chg sides, 2 chg coeffs, 1721 upgd conss, 22626 impls, 1905 clqs
(round 3, exhaustive) 0 del vars, 45 del conss, 10 add conss, 2 chg bounds, 0 chg sides, 2 chg coeffs, 1721 upgd conss, 22626 impls, 1905 clqs
presolving (4 rounds: 4 fast, 4 medium, 3 exhaustive):
0 deleted vars, 45 deleted constraints, 10 added constraints, 2 tightened bounds, 0 added holes, 0 changed sides, 2 changed coefficients
22626 implications, 1905 cliques
presolved problem has 26193 variables (17562 bin, 0 int, 0 impl, 8631 cont) and 22285 constraints
16893 constraints of type <varbound>
1599 constraints of type <setppc>
3345 constraints of type <linear>
420 constraints of type <logicor>
28 constraints of type <bounddisjunction>
transformed objective value is always integral (scale: 1)
Presolving Time: 28.20
transformed 100/100 original solutions to the transformed problem space
SCIP Status : solving was interrupted [time limit reached]
Solving Time (sec) : 301.66
Solving Nodes : 0 (total of 1049 nodes in 2 runs)
Primal Bound : +2.50779999999950e+04 (124 solutions)
Dual Bound : +2.30360421900002e+04
Gap : 8.86 %
-------------------------------------------------------------------------------
Summary
-------------------------------------------------------------------------------
(CVXPY) Dec 23 01:26:03 PM: Problem status: optimal_inaccurate
(CVXPY) Dec 23 01:26:03 PM: Optimal value: 2.508e+04
(CVXPY) Dec 23 01:26:03 PM: Compilation took 7.962e+01 seconds
(CVXPY) Dec 23 01:26:03 PM: Solver (including time spent in interface) took 3.072e+02 seconds
G_subdag = G_dag.edge_subgraph(P.expr.with_flow.value > 0.5)
G_dag.shape, G_subdag.shape
((12767, 38388), (12619, 25078))
rel_dag_compression = (1 - (G_subdag.num_edges / G_dag.num_edges)) * 100
print(f"KPNN edge compression (0-100%): {rel_dag_compression:.2f}%")
KPNN edge compression (0-100%): 34.67%
pruned_models, pruned_metrics = stratified_kfold(G_subdag, input_nn, outputs_pkn, file_weights=os.path.join(temp_weights, "pruned_weights"))
print("Validation metrics:")
for k, v in pruned_metrics.items():
print(f" - {k}: {np.mean(v):.3f}")
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 12778
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/pruned_weights_0.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 3s 75ms/step
> Fold 0 validation ROC-AUC=0.992
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 12778
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/pruned_weights_1.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 1s 73ms/step
> Fold 1 validation ROC-AUC=0.983
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 12778
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/pruned_weights_2.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 1s 69ms/step
> Fold 2 validation ROC-AUC=0.987
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 12778
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/pruned_weights_3.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 1s 68ms/step
> Fold 3 validation ROC-AUC=0.994
Building DAG NN model with CORNETO using Keras with JAX...
> N. inputs: 12459
> N. outputs: 1
> N. parameters: 12778
Compiling...
Fitting...
Weights saved to /var/folders/b4/gwkwsdb93sv11rtztqbm3l040000gn/T/tmpj5f_9545/pruned_weights_4.keras
11/11 ━━━━━━━━━━━━━━━━━━━━ 1s 68ms/step
> Fold 4 validation ROC-AUC=0.994
Validation metrics:
- accuracy: 0.961
- precision: 0.971
- recall: 0.949
- f1: 0.960
- roc_auc: 0.990
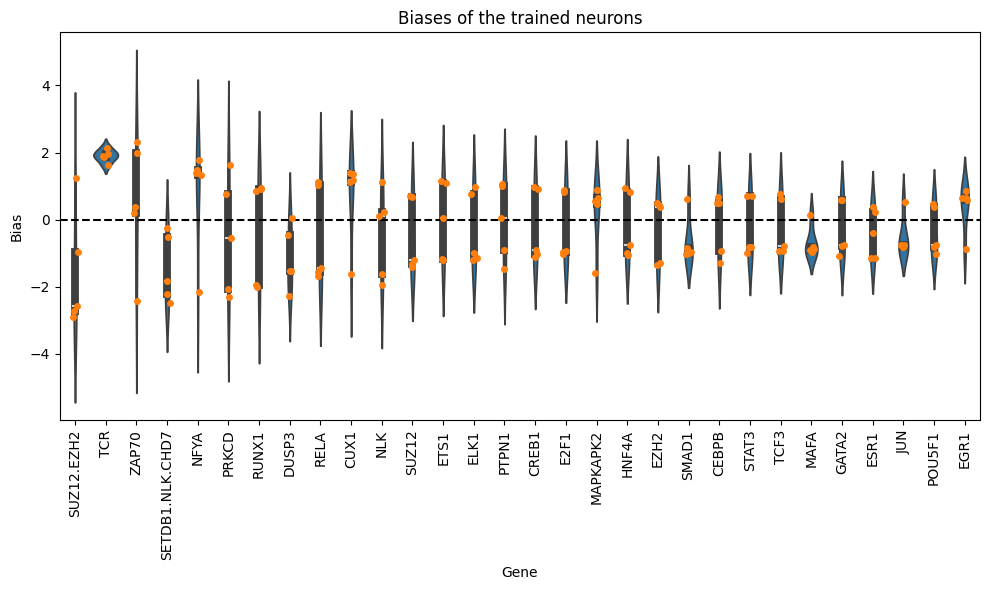
df_biases_pruned = load_biases(file=os.path.join(temp_weights, "pruned_weights"), folds=5)
df_biases_pruned.sort_values(by="abs_bias", ascending=False).head(10)
| bias | abs_bias | pow2_bias | ||
|---|---|---|---|---|
| fold | gene | |||
| 1 | SUZ12.EZH2 | -2.892245 | 2.892245 | 8.365083 |
| 4 | SUZ12.EZH2 | -2.736529 | 2.736529 | 7.488593 |
| 2 | SUZ12.EZH2 | -2.575348 | 2.575348 | 6.632418 |
| 0 | SETDB1.NLK.CHD7 | -2.476090 | 2.476090 | 6.131024 |
| 4 | ZAP70 | -2.432173 | 2.432173 | 5.915468 |
| 3 | ZAP70 | 2.325642 | 2.325642 | 5.408611 |
| 0 | PRKCD | -2.315197 | 2.315197 | 5.360137 |
| DUSP3 | -2.269532 | 2.269532 | 5.150774 | |
| 4 | SETDB1.NLK.CHD7 | -2.219929 | 2.219929 | 4.928085 |
| 3 | NFYA | -2.163613 | 2.163613 | 4.681221 |
df_biases_prunedr = df_biases_pruned.copy().reset_index()
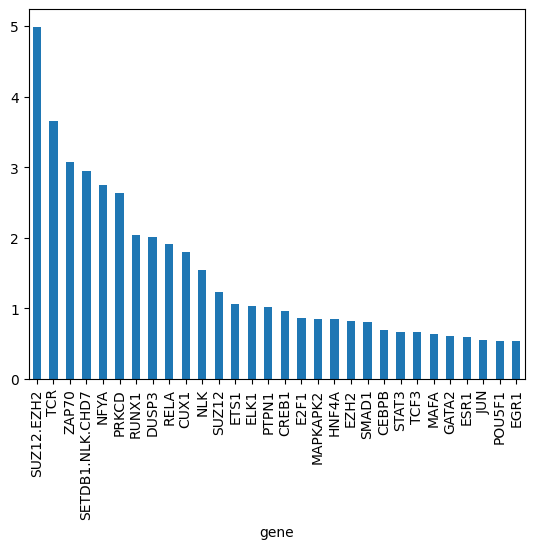
gene_biases_score = df_biases_prunedr.groupby("gene")["pow2_bias"].mean().sort_values(ascending=False)
gene_biases_score
gene
SUZ12.EZH2 4.990975
TCR 3.651254
ZAP70 3.080308
SETDB1.NLK.CHD7 2.942616
NFYA 2.748323
...
PBX1 0.007306
NR2F1 0.005788
ASH2L 0.002699
BCL3 0.002370
GATA3 0.000018
Name: pow2_bias, Length: 160, dtype: float32
gene_biases_score.head(30).plot.bar()
<Axes: xlabel='gene'>
# Get the top genes sorted by mean bias
top_genes = gene_biases_score.head(30).index
# Filter the DataFrame to include only rows with these top genes
filtered_df = df_biases_prunedr[df_biases_prunedr['gene'].isin(top_genes)]
plt.figure(figsize=(10, 6))
sns.violinplot(data=filtered_df, x="gene", y="bias", order=top_genes, dodge=True)
sns.stripplot(data=filtered_df, x="gene", y="bias", order=top_genes, dodge=True)
plt.xticks(rotation=90)
plt.title('Biases of the trained neurons')
plt.xlabel('Gene')
plt.ylabel('Bias')
plt.axhline(0, linestyle="--", color="k")
plt.tight_layout()
param_compression = (1-(pruned_models[0].count_params()/models[0].count_params())) * 100
print(f"Parameter compression: {param_compression:.2f}%")
Parameter compression: 51.30%
perf_degradation = ((np.mean(metrics["roc_auc"]) - np.mean(pruned_metrics["roc_auc"])) / np.mean(metrics["roc_auc"])) * 100
print(f"Degradation in ROC-AUC after compression (positive = decrease in performance, negative = increase in performance): {perf_degradation:.2f}%")
Degradation in ROC-AUC after compression (positive = decrease in performance, negative = increase in performance): -0.04%
