Implementing an alternative network sampler#
In this tutorial, we demonstrate how to use CORNETO to implement a simple sampler to explore alternative solutions for network inference problems. By perturbing the objective function, we can explore the solution space and identify robust as well as variable components of the network.
We will illustrate this process using a simple example of a network inference problem with CARNIVAL, using the data from the CARNIVAL transcriptomics tutorial.
NOTE: This notebook uses
gurobias the solver andcvxpyas the backend to accelerate the generation of alternative solutions using thewarm_startoption from cvxpy
import corneto as cn
cn.info()
|
|
|
from corneto._data import GraphData
# Load dataset from tutorial
dataset = GraphData.load("data/carnival_transcriptomics_dataset.zip")
from corneto.methods.future import CarnivalFlow
m = CarnivalFlow(lambda_reg=0.1)
P = m.build(dataset.graph, dataset.data)
P.objectives
Unreachable vertices for sample: 0
[error_sample1_0: Expression(AFFINE, UNKNOWN, ()),
multi_sample_regularization_edge_has_signal: Expression(AFFINE, NONNEGATIVE, ())]
# We will sample solutions by perturbing the edge_has_signal variable
P.expr
{'edge_inhibits': edge_inhibits: Variable((3374, 1), edge_inhibits, boolean=True),
'_dag_layer': _dag_layer: Variable((970, 1), _dag_layer),
'_flow': _flow: Variable((3374,), _flow),
'edge_activates': edge_activates: Variable((3374, 1), edge_activates, boolean=True),
'const0x16b856374e4a9f30': const0x16b856374e4a9f30: Constant(CONSTANT, NONNEGATIVE, (970, 3374)),
'const0x72fe108f440b0c6': const0x72fe108f440b0c6: Constant(CONSTANT, NONNEGATIVE, (970, 3374)),
'flow': _flow: Variable((3374,), _flow),
'vertex_value': Expression(AFFINE, UNKNOWN, (970, 1)),
'vertex_activated': Expression(AFFINE, NONNEGATIVE, (970, 1)),
'vertex_inhibited': Expression(AFFINE, NONNEGATIVE, (970, 1)),
'edge_value': Expression(AFFINE, UNKNOWN, (3374, 1)),
'edge_has_signal': Expression(AFFINE, NONNEGATIVE, (3374, 1))}
Sampling Solutions Using Random Perturbation#
We’ll implement a generic approach to sample alternative solutions by:
Introducing small random perturbations to the objective function
Re-optimizing the model with these perturbations
Collecting solutions that remain close to optimal (within a tolerance threshold)
This technique allows us to explore the space of near-optimal solutions and assess the robustness of different model components.
import numpy as np
import pandas as pd
def sample_alternative_solutions(
problem,
target_variable,
percentage=0.1,
scale=0.03,
rel_opt_tol=0.05,
max_samples=30,
time_limit=60,
):
"""Sample alternative solutions by perturbing the objective function.
Parameters:
-----------
problem : CORNETO optimization problem
The optimization problem to sample
target_variable : variable expression
The variable to perturb (e.g., P.expr.edge_has_signal or P.expr.vertex_value)
percentage : float
Percentage of elements to perturb
scale : float
Scale parameter for normal distribution perturbation
rel_opt_tol : float
Relative optimality tolerance
max_samples : int
Maximum number of samples to generate
time_limit : int
Time limit for solver
Returns:
--------
dict : A dictionary containing the results
"""
# Save the original problem state
original_problem = problem.solve(solver="gurobi", verbosity=1)
# Store original objective values
orig_objectives = []
for o in problem.objectives:
print(o.name, o.value)
orig_objectives.append(o.value)
# Prepare for perturbation
shape = target_variable.shape
num_elements = shape[0]
n_elements = int(num_elements * percentage)
vec = np.zeros(shape=num_elements)
pert = problem.backend.Parameter("perturbation", shape=vec.shape, value=vec)
# Add perturbation objective
problem.add_objective(target_variable.T @ pert, name="perturbation")
# Sample solutions
selected_edges = []
edge_values = []
vertex_values = []
for i in range(max_samples):
# Generate random perturbation
c = np.random.normal(scale=scale, size=n_elements)
random_indices = np.random.choice(vec.shape[0], size=n_elements, replace=False)
vec[:] = 0 # Reset vector
vec[random_indices] = c
# Update parameter and solve
pert.value = vec
solved_problem = problem.solve(
solver="gurobi",
warm_start=True,
ignore_dpp=True,
TimeLimit=time_limit,
verbosity=0,
)
print(
f"Sample {i + 1}/{max_samples}: Objective value = {solved_problem.value:.4f}"
)
# Check if solution is within tolerance
accept = True
for o_orig, o in zip(orig_objectives, problem.objectives):
if o.name == "perturbation":
continue
relative_error = np.abs(o.value - o_orig) / np.abs(o_orig + 1e-10)
print(f" {o.name}: {o.value:.4f}, rel error: {relative_error:.4f}")
if relative_error > rel_opt_tol:
print(f" > Rejected! ({relative_error:.4f} >= {rel_opt_tol})")
accept = False
break
if accept:
selected_edges.append(problem.expr.edge_has_signal.value > 0.5)
edge_values.append(problem.expr.edge_value.value)
vertex_values.append(problem.expr.vertex_value.value)
return {
"selected_edges": selected_edges,
"edge_values": edge_values,
"vertex_values": vertex_values,
}
Edge-Based Perturbation#
First, we’ll sample solutions by perturbing the edge variables. This allows us to explore alternative network topologies where different edges might be selected.
# Create a fresh model
m = CarnivalFlow(lambda_reg=0.1)
P = m.build(dataset.graph, dataset.data)
# Sample solutions by perturbing edge_has_signal
edge_results = sample_alternative_solutions(
problem=P,
target_variable=P.expr.edge_has_signal,
percentage=0.10,
scale=0.03,
rel_opt_tol=0.05,
max_samples=30,
)
# Analyze edge-based results
df_sols = pd.DataFrame(
np.concatenate(edge_results["selected_edges"], axis=1), index=m.processed_graph.E
).astype(int)
print(f"Generated {df_sols.shape[1]} alternative solutions")
df_sols.head()
Unreachable vertices for sample: 0
===============================================================================
CVXPY
v1.6.4
===============================================================================
(CVXPY) Apr 10 07:10:17 PM: Your problem has 11092 variables, 31652 constraints, and 1 parameters.
(CVXPY) Apr 10 07:10:17 PM: It is compliant with the following grammars: DCP, DQCP
(CVXPY) Apr 10 07:10:17 PM: CVXPY will first compile your problem; then, it will invoke a numerical solver to obtain a solution.
(CVXPY) Apr 10 07:10:17 PM: Your problem is compiled with the CPP canonicalization backend.
-------------------------------------------------------------------------------
Compilation
-------------------------------------------------------------------------------
(CVXPY) Apr 10 07:10:17 PM: Compiling problem (target solver=GUROBI).
(CVXPY) Apr 10 07:10:17 PM: Reduction chain: CvxAttr2Constr -> Qp2SymbolicQp -> QpMatrixStuffing -> GUROBI
(CVXPY) Apr 10 07:10:17 PM: Applying reduction CvxAttr2Constr
(CVXPY) Apr 10 07:10:17 PM: Applying reduction Qp2SymbolicQp
(CVXPY) Apr 10 07:10:17 PM: Applying reduction QpMatrixStuffing
(CVXPY) Apr 10 07:10:17 PM: Applying reduction GUROBI
(CVXPY) Apr 10 07:10:17 PM: Finished problem compilation (took 5.276e-02 seconds).
(CVXPY) Apr 10 07:10:17 PM: (Subsequent compilations of this problem, using the same arguments, should take less time.)
-------------------------------------------------------------------------------
Numerical solver
-------------------------------------------------------------------------------
(CVXPY) Apr 10 07:10:17 PM: Invoking solver GUROBI to obtain a solution.
Set parameter Username
Set parameter LicenseID to value 2593994
Academic license - for non-commercial use only - expires 2025-12-02
Set parameter OutputFlag to value 1
Set parameter QCPDual to value 1
Gurobi Optimizer version 12.0.1 build v12.0.1rc0 (mac64[arm] - Darwin 24.4.0 24E248)
CPU model: Apple M4 Pro
Thread count: 12 physical cores, 12 logical processors, using up to 12 threads
Non-default parameters:
QCPDual 1
Optimize a model with 31652 rows, 11092 columns and 111482 nonzeros
Model fingerprint: 0xca949f9c
Variable types: 4344 continuous, 6748 integer (6748 binary)
Coefficient statistics:
Matrix range [1e+00, 1e+03]
Objective range [1e-01, 8e+00]
Bounds range [1e+00, 1e+00]
RHS range [1e+00, 1e+03]
Found heuristic solution: objective 0.0000000
Presolve removed 18549 rows and 1627 columns
Presolve time: 0.14s
Presolved: 13103 rows, 9465 columns, 82774 nonzeros
Variable types: 3602 continuous, 5863 integer (5853 binary)
Root relaxation: objective -2.060081e+02, 1253 iterations, 0.02 seconds (0.06 work units)
Nodes | Current Node | Objective Bounds | Work
Expl Unexpl | Obj Depth IntInf | Incumbent BestBd Gap | It/Node Time
0 0 -206.00811 0 69 0.00000 -206.00811 - - 0s
0 0 -202.60774 0 71 0.00000 -202.60774 - - 0s
0 0 -202.32031 0 65 0.00000 -202.32031 - - 0s
0 0 -202.32031 0 60 0.00000 -202.32031 - - 0s
0 0 -202.27031 0 59 0.00000 -202.27031 - - 0s
0 0 -202.27031 0 47 0.00000 -202.27031 - - 0s
0 0 -202.27031 0 51 0.00000 -202.27031 - - 0s
0 0 -202.27031 0 51 0.00000 -202.27031 - - 0s
0 0 -202.27031 0 14 0.00000 -202.27031 - - 0s
0 0 -202.27031 0 45 0.00000 -202.27031 - - 0s
0 0 -202.25364 0 34 0.00000 -202.25364 - - 0s
0 0 -202.23697 0 55 0.00000 -202.23697 - - 0s
H 0 0 -198.0703069 -202.23697 2.10% - 0s
H 0 0 -198.9703069 -202.22031 1.63% - 0s
0 0 -202.22031 0 35 -198.97031 -202.22031 1.63% - 0s
0 0 -202.20364 0 35 -198.97031 -202.20364 1.63% - 0s
0 0 -202.15364 0 65 -198.97031 -202.15364 1.60% - 0s
H 0 0 -201.7703069 -202.15364 0.19% - 0s
0 0 -202.15364 0 67 -201.77031 -202.15364 0.19% - 0s
0 0 -202.10364 0 70 -201.77031 -202.10364 0.17% - 0s
0 0 -202.10364 0 68 -201.77031 -202.10364 0.17% - 0s
0 0 -202.09531 0 58 -201.77031 -202.09531 0.16% - 0s
0 0 -202.07031 0 57 -201.77031 -202.07031 0.15% - 0s
0 0 -202.07031 0 51 -201.77031 -202.07031 0.15% - 0s
0 0 -202.07031 0 53 -201.77031 -202.07031 0.15% - 0s
0 0 -202.07031 0 24 -201.77031 -202.07031 0.15% - 0s
0 0 -202.07031 0 26 -201.77031 -202.07031 0.15% - 0s
0 0 -202.07031 0 25 -201.77031 -202.07031 0.15% - 0s
0 0 -202.07031 0 25 -201.77031 -202.07031 0.15% - 1s
0 0 -202.07031 0 37 -201.77031 -202.07031 0.15% - 1s
0 0 -202.07031 0 37 -201.77031 -202.07031 0.15% - 1s
0 0 -202.07031 0 37 -201.77031 -202.07031 0.15% - 1s
0 0 -202.07031 0 37 -201.77031 -202.07031 0.15% - 1s
0 2 -202.07031 0 37 -201.77031 -202.07031 0.15% - 1s
H 28 16 -201.8703069 -201.97031 0.05% 22.7 1s
H 76 38 -201.9703069 -201.97031 0.00% 17.7 1s
Cutting planes:
Learned: 2
Gomory: 5
Cover: 1
Implied bound: 26
Clique: 2
MIR: 37
Inf proof: 1
Zero half: 7
Mod-K: 2
RLT: 11
Relax-and-lift: 6
Explored 93 nodes (8523 simplex iterations) in 1.37 seconds (2.08 work units)
Thread count was 12 (of 12 available processors)
Solution count 6: -201.97 -201.87 -201.77 ... 0
No other solutions better than -201.97
Optimal solution found (tolerance 1.00e-04)
Best objective -2.019703069210e+02, best bound -2.019703069210e+02, gap 0.0000%
-------------------------------------------------------------------------------
Summary
-------------------------------------------------------------------------------
(CVXPY) Apr 10 07:10:19 PM: Problem status: optimal
(CVXPY) Apr 10 07:10:19 PM: Optimal value: 1.622e+01
(CVXPY) Apr 10 07:10:19 PM: Compilation took 5.276e-02 seconds
(CVXPY) Apr 10 07:10:19 PM: Solver (including time spent in interface) took 1.406e+00 seconds
error_sample1_0 7.0169713497161865
multi_sample_regularization_edge_has_signal 92.0
Sample 1/30: Objective value = 15.9878
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 2/30: Objective value = 16.2013
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 3/30: Objective value = 16.2013
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 4/30: Objective value = 16.1317
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 5/30: Objective value = 16.1711
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 6/30: Objective value = 16.2996
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 93.0000, rel error: 0.0109
Sample 7/30: Objective value = 16.2228
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 8/30: Objective value = 16.2558
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 9/30: Objective value = 16.1316
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 10/30: Objective value = 15.8947
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 11/30: Objective value = 16.0179
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 12/30: Objective value = 15.8665
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 13/30: Objective value = 16.1839
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 14/30: Objective value = 16.2407
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 15/30: Objective value = 16.1211
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 16/30: Objective value = 16.0665
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 17/30: Objective value = 15.8804
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 18/30: Objective value = 16.2178
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 19/30: Objective value = 16.0880
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 20/30: Objective value = 16.2337
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 21/30: Objective value = 16.2322
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 22/30: Objective value = 15.8936
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 23/30: Objective value = 16.1274
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 24/30: Objective value = 16.0623
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 25/30: Objective value = 15.9696
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 93.0000, rel error: 0.0109
Sample 26/30: Objective value = 16.0228
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 27/30: Objective value = 15.9609
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 28/30: Objective value = 16.1027
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 29/30: Objective value = 16.2100
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 30/30: Objective value = 16.1709
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Generated 30 alternative solutions
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (SMAD3) | (MYOD1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (GRK2) | (BDKRB2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (MAPK14) | (MAPKAPK2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (DEPTOR_EEF1A1_MLST8_MTOR_PRR5_RICTOR) | (FBXW8) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (SLK) | (MAP3K5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 30 columns
# Analyze variability in edge selection
df_var = pd.concat([df_sols.mean(axis=1), df_sols.std(axis=1)], axis=1)
df_var.columns = ["mean", "std"]
print("Edges with highest variability across solutions:")
df_var.sort_values(by="std", ascending=False).head(50).sort_values(by="mean")
Edges with highest variability across solutions:
| mean | std | ||
|---|---|---|---|
| (PPP2CA) | (RB1) | 0.300000 | 0.466092 |
| (PML) | (SMAD3) | 0.300000 | 0.466092 |
| (MAPK1) | (JUN) | 0.300000 | 0.466092 |
| (GSK3B) | (NFKB1_RELA) | 0.300000 | 0.466092 |
| (MAPK3) | (PML) | 0.333333 | 0.479463 |
| (CTNNB1) | (KLF4) | 0.333333 | 0.479463 |
| (PHLPP1) | (STK4) | 0.333333 | 0.479463 |
| (GSK3B) | (CDKN1A) | 0.366667 | 0.490133 |
| (MAPK3) | (GSK3B) | 0.366667 | 0.490133 |
| (APC_AXIN1_GSK3B) | (ROR2) | 0.400000 | 0.498273 |
| (RARB) | (RXRB) | 0.400000 | 0.498273 |
| (ROR2) | (MAPK8) | 0.400000 | 0.498273 |
| (THRA) | (RARB) | 0.400000 | 0.498273 |
| (PRKCD) | (STAT1) | 0.400000 | 0.498273 |
| (CDK1) | (HMGA1) | 0.400000 | 0.498273 |
| (MAP2K1) | 0.400000 | 0.498273 | |
| (WWTR1) | (NKX2-1) | 0.400000 | 0.498273 |
| (MAP2K1) | (MAPK3) | 0.400000 | 0.498273 |
| (ABL1) | (CEBPB) | 0.433333 | 0.504007 |
| (NFKB1_RELA) | (EGR1) | 0.466667 | 0.507416 |
| (SMAD2) | (MEF2A) | 0.466667 | 0.507416 |
| (TP53) | (ETS1) | 0.500000 | 0.508548 |
| (MAPK3) | (ETS1) | 0.500000 | 0.508548 |
| (RELA) | (EGR1) | 0.533333 | 0.507416 |
| (MAPK14) | (MEF2A) | 0.533333 | 0.507416 |
| (HIPK2) | (HMGA1) | 0.533333 | 0.507416 |
| (JUN) | (SPI1) | 0.533333 | 0.507416 |
| (HIPK2) | (TP53) | 0.566667 | 0.504007 |
| (SMAD3) | (CEBPB) | 0.566667 | 0.504007 |
| (PPP2CA) | (PRKCD) | 0.566667 | 0.504007 |
| (THRA) | (RARG) | 0.600000 | 0.498273 |
| (PRKCD) | (MAPK8) | 0.600000 | 0.498273 |
| (RARG) | (RXRB) | 0.600000 | 0.498273 |
| (SMAD3) | (NKX2-1) | 0.600000 | 0.498273 |
| (CDC25A) | (MAPK3) | 0.600000 | 0.498273 |
| (MAPK8) | (STAT1) | 0.600000 | 0.498273 |
| (CDK1) | (CDC25A) | 0.600000 | 0.498273 |
| (MAPK14) | (GSK3B) | 0.633333 | 0.490133 |
| (CDKN1A) | 0.633333 | 0.490133 | |
| (MAPK1) | (APC_AXIN1_GSK3B) | 0.666667 | 0.479463 |
| (CSNK1D) | (WWTR1) | 0.666667 | 0.479463 |
| (APC_AXIN1_GSK3B) | (CSNK1D) | 0.666667 | 0.479463 |
| (MUC1) | (KLF4) | 0.666667 | 0.479463 |
| (GSK3B) | (MUC1) | 0.666667 | 0.479463 |
| (CDK1) | (PML) | 0.666667 | 0.479463 |
| (TBK1) | (IRF3) | 0.666667 | 0.479463 |
| (MAPK14) | (SMAD3) | 0.700000 | 0.466092 |
| (SMAD3) | (SMAD4) | 0.700000 | 0.466092 |
| (ABL1) | (RB1) | 0.700000 | 0.466092 |
| (PHLPP1) | (PRKCA) | 0.733333 | 0.449776 |
# Analyze edge values across solutions
df_edge_val = pd.DataFrame(
np.concatenate(edge_results["edge_values"], axis=1), index=m.processed_graph.E
).astype(int)
df_edge_val.head()
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (SMAD3) | (MYOD1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (GRK2) | (BDKRB2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (MAPK14) | (MAPKAPK2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (DEPTOR_EEF1A1_MLST8_MTOR_PRR5_RICTOR) | (FBXW8) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (SLK) | (MAP3K5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 30 columns
Vertex-Based Perturbation#
Next, we’ll sample solutions by perturbing vertex variables. This approach can highlight alternative active node sets that are consistent with the data.
# Create a fresh model
m = CarnivalFlow(lambda_reg=0.1)
P = m.build(dataset.graph, dataset.data)
# Sample solutions by perturbing vertex_value
vertex_results = sample_alternative_solutions(
problem=P,
target_variable=P.expr.vertex_value,
percentage=0.10,
scale=0.03,
rel_opt_tol=0.05,
max_samples=30,
)
# Analyze vertex-based results
df_vertex_val = pd.DataFrame(
np.concatenate(vertex_results["vertex_values"], axis=1), index=m.processed_graph.V
).astype(int)
print(f"Generated {df_vertex_val.shape[1]} alternative solutions")
df_vertex_val.head()
Unreachable vertices for sample: 0
===============================================================================
CVXPY
v1.6.4
===============================================================================
(CVXPY) Apr 10 07:11:09 PM: Your problem has 11092 variables, 31652 constraints, and 1 parameters.
(CVXPY) Apr 10 07:11:09 PM: It is compliant with the following grammars: DCP, DQCP
(CVXPY) Apr 10 07:11:09 PM: CVXPY will first compile your problem; then, it will invoke a numerical solver to obtain a solution.
(CVXPY) Apr 10 07:11:09 PM: Your problem is compiled with the CPP canonicalization backend.
-------------------------------------------------------------------------------
Compilation
-------------------------------------------------------------------------------
(CVXPY) Apr 10 07:11:09 PM: Compiling problem (target solver=GUROBI).
(CVXPY) Apr 10 07:11:09 PM: Reduction chain: CvxAttr2Constr -> Qp2SymbolicQp -> QpMatrixStuffing -> GUROBI
(CVXPY) Apr 10 07:11:09 PM: Applying reduction CvxAttr2Constr
(CVXPY) Apr 10 07:11:09 PM: Applying reduction Qp2SymbolicQp
(CVXPY) Apr 10 07:11:09 PM: Applying reduction QpMatrixStuffing
(CVXPY) Apr 10 07:11:09 PM: Applying reduction GUROBI
(CVXPY) Apr 10 07:11:09 PM: Finished problem compilation (took 5.829e-02 seconds).
(CVXPY) Apr 10 07:11:09 PM: (Subsequent compilations of this problem, using the same arguments, should take less time.)
-------------------------------------------------------------------------------
Numerical solver
-------------------------------------------------------------------------------
(CVXPY) Apr 10 07:11:09 PM: Invoking solver GUROBI to obtain a solution.
Set parameter OutputFlag to value 1
Set parameter QCPDual to value 1
Gurobi Optimizer version 12.0.1 build v12.0.1rc0 (mac64[arm] - Darwin 24.4.0 24E248)
CPU model: Apple M4 Pro
Thread count: 12 physical cores, 12 logical processors, using up to 12 threads
Non-default parameters:
QCPDual 1
Optimize a model with 31652 rows, 11092 columns and 111482 nonzeros
Model fingerprint: 0x926525f9
Variable types: 4344 continuous, 6748 integer (6748 binary)
Coefficient statistics:
Matrix range [1e+00, 1e+03]
Objective range [1e-01, 8e+00]
Bounds range [1e+00, 1e+00]
RHS range [1e+00, 1e+03]
Found heuristic solution: objective 0.0000000
Presolve removed 18549 rows and 1627 columns
Presolve time: 0.15s
Presolved: 13103 rows, 9465 columns, 82774 nonzeros
Variable types: 3602 continuous, 5863 integer (5853 binary)
Root relaxation: objective -2.060081e+02, 1119 iterations, 0.02 seconds (0.05 work units)
Nodes | Current Node | Objective Bounds | Work
Expl Unexpl | Obj Depth IntInf | Incumbent BestBd Gap | It/Node Time
0 0 -206.00811 0 64 0.00000 -206.00811 - - 0s
0 0 -202.60774 0 66 0.00000 -202.60774 - - 0s
H 0 0 -121.0727864 -202.45357 67.2% - 0s
H 0 0 -125.9042639 -202.45357 60.8% - 0s
0 0 -202.32031 0 64 -125.90426 -202.32031 60.7% - 0s
0 0 -202.27031 0 65 -125.90426 -202.27031 60.7% - 0s
0 0 -202.27031 0 54 -125.90426 -202.27031 60.7% - 0s
0 0 -202.27031 0 52 -125.90426 -202.27031 60.7% - 0s
H 0 0 -131.7216934 -202.27031 53.6% - 0s
0 0 -202.27031 0 56 -131.72169 -202.27031 53.6% - 0s
0 0 -202.27031 0 42 -131.72169 -202.27031 53.6% - 0s
0 0 -202.27031 0 40 -131.72169 -202.27031 53.6% - 0s
H 0 0 -134.7525108 -202.27031 50.1% - 0s
0 0 -202.25020 0 28 -134.75251 -202.25020 50.1% - 0s
H 0 0 -140.2462506 -202.22020 44.2% - 0s
0 0 -202.22020 0 28 -140.24625 -202.22020 44.2% - 0s
0 0 -202.22020 0 18 -140.24625 -202.22020 44.2% - 0s
H 0 0 -199.6703069 -202.20354 1.27% - 0s
H 0 0 -200.1703069 -202.20354 1.02% - 0s
0 0 -202.20354 0 18 -200.17031 -202.20354 1.02% - 0s
H 0 0 -201.9703069 -202.13697 0.08% - 0s
0 0 -202.13697 0 22 -201.97031 -202.13697 0.08% - 0s
0 0 -202.13697 0 22 -201.97031 -202.13697 0.08% - 0s
0 0 -202.07031 0 2 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 2 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 2 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 5 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 5 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 25 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 29 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 29 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 22 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 26 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 26 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 23 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 26 -201.97031 -202.07031 0.05% - 0s
0 0 -202.07031 0 6 -201.97031 -202.07031 0.05% - 0s
Cutting planes:
Learned: 2
Gomory: 4
Cover: 1
Implied bound: 11
Clique: 4
MIR: 9
Zero half: 7
Mod-K: 1
RLT: 10
Relax-and-lift: 1
BQP: 1
PSD: 2
Explored 1 nodes (4497 simplex iterations) in 0.86 seconds (1.20 work units)
Thread count was 12 (of 12 available processors)
Solution count 9: -201.97 -200.17 -199.67 ... 0
No other solutions better than -201.97
Optimal solution found (tolerance 1.00e-04)
Best objective -2.019703069210e+02, best bound -2.019703069210e+02, gap 0.0000%
-------------------------------------------------------------------------------
Summary
-------------------------------------------------------------------------------
(CVXPY) Apr 10 07:11:10 PM: Problem status: optimal
(CVXPY) Apr 10 07:11:10 PM: Optimal value: 1.622e+01
(CVXPY) Apr 10 07:11:10 PM: Compilation took 5.829e-02 seconds
(CVXPY) Apr 10 07:11:10 PM: Solver (including time spent in interface) took 8.972e-01 seconds
error_sample1_0 7.0169713497161865
multi_sample_regularization_edge_has_signal 92.0
Sample 1/30: Objective value = 16.2117
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 2/30: Objective value = 16.2916
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 93.0000, rel error: 0.0109
Sample 3/30: Objective value = 16.2261
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 4/30: Objective value = 16.2643
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 5/30: Objective value = 16.3308
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 6/30: Objective value = 16.2693
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 7/30: Objective value = 16.3319
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 8/30: Objective value = 16.1480
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 9/30: Objective value = 16.3680
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 10/30: Objective value = 16.1878
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 11/30: Objective value = 16.0145
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 12/30: Objective value = 16.0929
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 13/30: Objective value = 16.3187
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 14/30: Objective value = 16.1250
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 15/30: Objective value = 16.1789
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 16/30: Objective value = 16.2809
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 17/30: Objective value = 16.3439
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 18/30: Objective value = 16.1387
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 19/30: Objective value = 16.1296
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 20/30: Objective value = 16.1100
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 21/30: Objective value = 16.2697
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 22/30: Objective value = 16.1848
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 23/30: Objective value = 16.0936
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 24/30: Objective value = 16.1869
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 25/30: Objective value = 16.2711
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 26/30: Objective value = 16.1187
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 27/30: Objective value = 16.2951
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 28/30: Objective value = 16.2525
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 29/30: Objective value = 16.2769
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Sample 30/30: Objective value = 16.1914
error_sample1_0: 7.0170, rel error: 0.0000
multi_sample_regularization_edge_has_signal: 92.0000, rel error: 0.0000
Generated 30 alternative solutions
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DIAPH1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PRKD1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ALDH2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MDM2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RAD21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 30 columns
# Focus on signaling proteins with no measurement data
df_sig_prot_pred = df_vertex_val.loc[
df_vertex_val.index.difference(dataset.data.query.pluck_features())
]
df_sig_prot_pred.head()
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAK1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABI1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABL1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | ... | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 |
| ABL2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ABRAXAS1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 30 columns
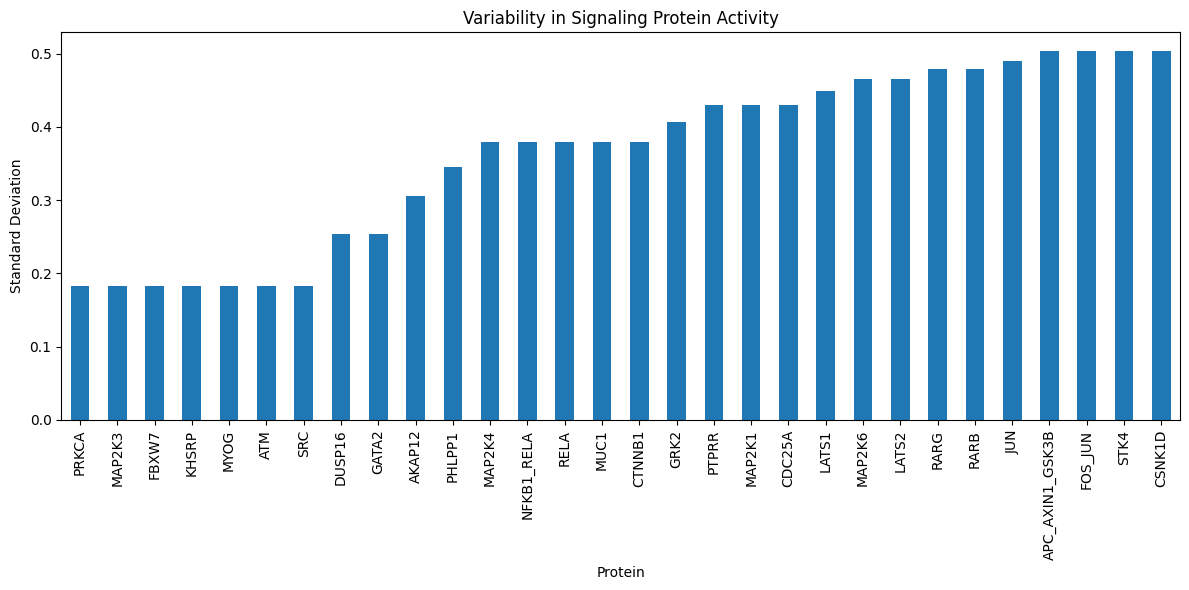
# Visualize the variability in predicted signaling protein activities
import matplotlib.pyplot as plt
plt.figure(figsize=(12, 6))
df_sig_prot_pred.std(axis=1).sort_values().tail(30).plot.bar(
title="Variability in Signaling Protein Activity"
)
plt.xlabel("Protein")
plt.ylabel("Standard Deviation")
plt.tight_layout()
plt.show()
Summary#
In this notebook, we:
Created a generic sampling method for exploring alternative network solutions
Applied the method to both edge and vertex perturbations
Analyzed the variability in solutions to identify:
Edges with high uncertainty (appearing in some but not all solutions)
Signaling proteins with variable activity across solutions
This approach helps assess the robustness of network inference results and can guide further experimental validation efforts by highlighting the most uncertain parts of the network.
