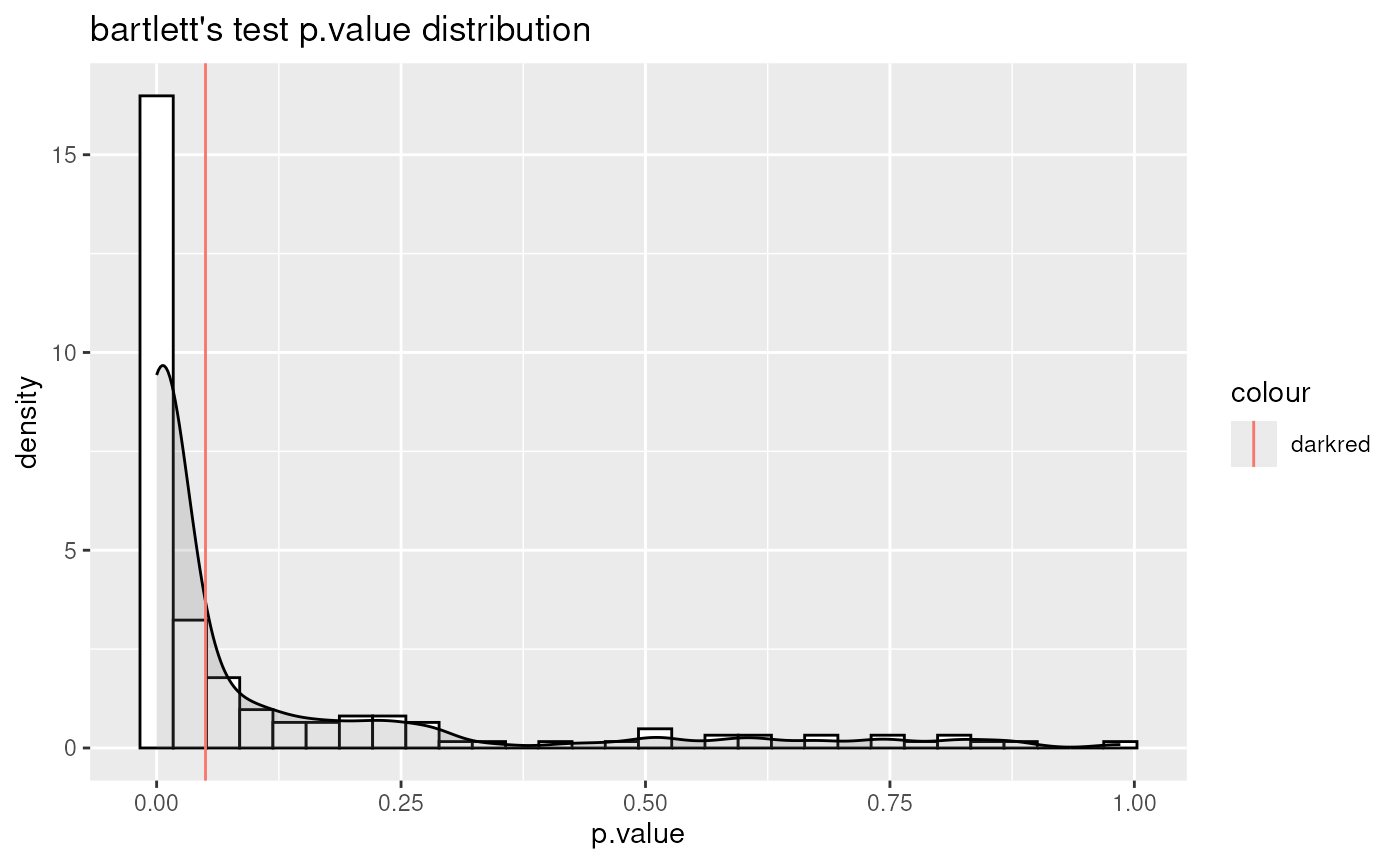
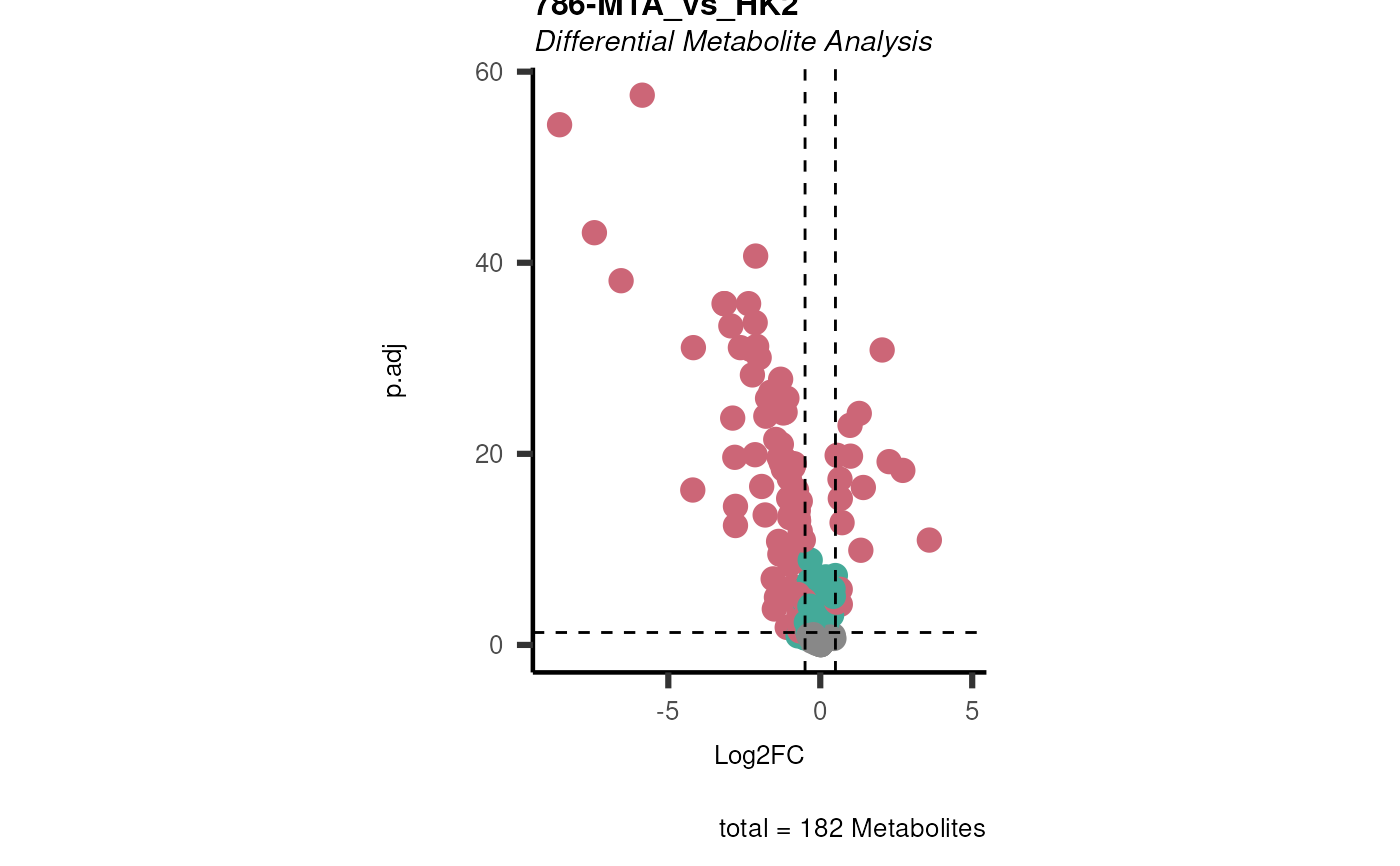
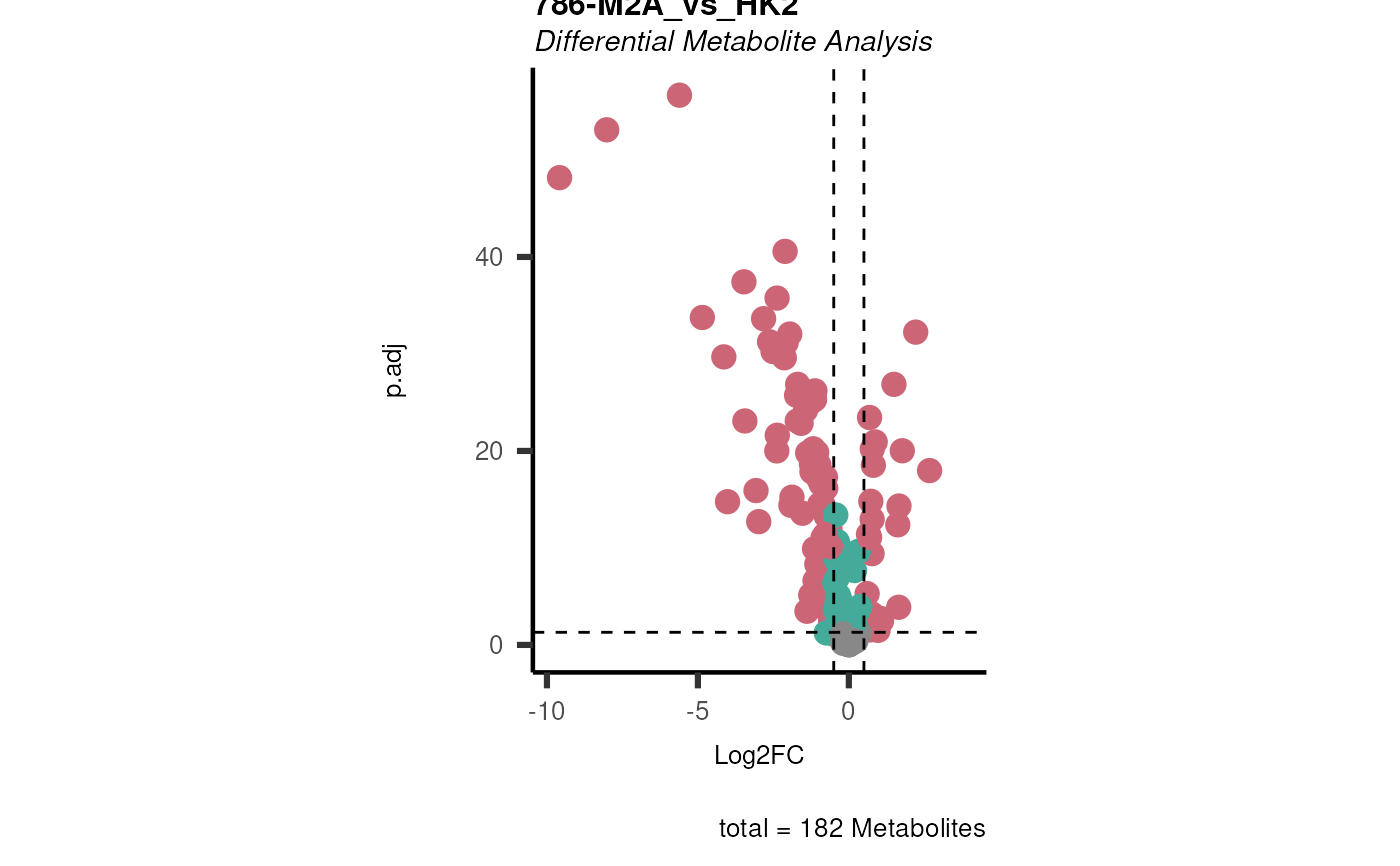
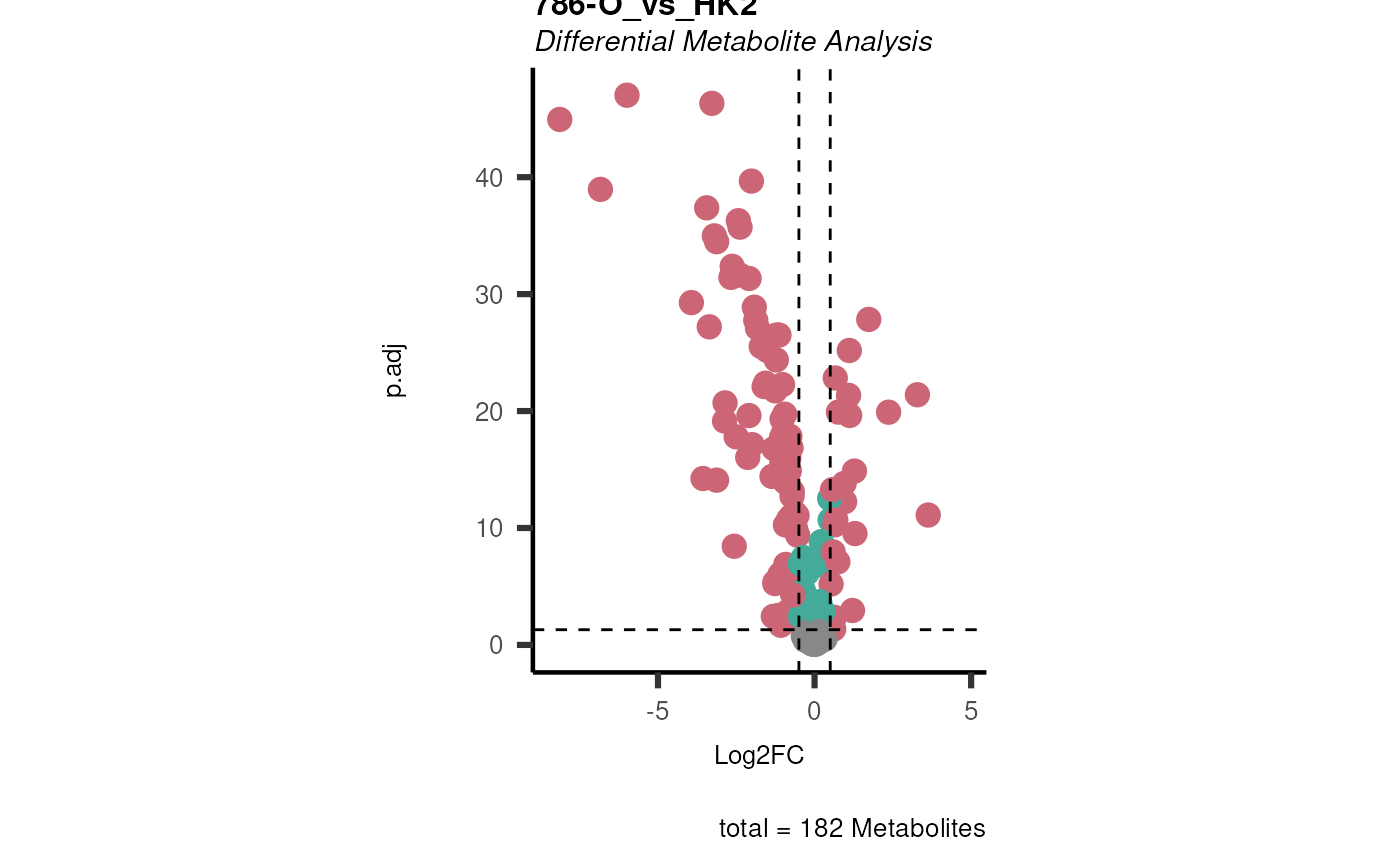
Performs differential metabolite analysis to obtain Log2FC, p-value, adjusted p-value, and t-value when comparing two or multiple conditions.
Usage
dma(
data,
metadata_sample = NULL,
metadata_info = c(Conditions = "Conditions", Numerator = NULL, Denominator = NULL),
pval = "lmFit",
padj = "fdr",
metadata_feature = NULL,
core = FALSE,
vst = FALSE,
shapiro = TRUE,
bartlett = TRUE,
transform = TRUE,
save_plot = "svg",
save_table = "csv",
print_plot = TRUE,
path = NULL
)Arguments
- data
SummarizedExperiment or data frame. If SummarizedExperiment, metadata_sample is extracted from colData and metadata_feature from rowData. If data frame, provide unique sample identifiers as row names and metabolite numerical values in columns with metabolite identifiers as column names. Use NA for undetected metabolites.
- metadata_sample
Data frame (optional). Only required if data is not a SummarizedExperiment. Contains metadata information about samples, combined with input data based on unique sample identifiers used as row names. Default: NULL.
- metadata_info
Named character vector (optional). Includes conditions column information on numerator or denominator: c(Conditions="ColumnName", Numerator="ColumnName", Denominator="ColumnName"). Denominator and Numerator specify which comparisons are performed (one-vs-one, all-vs-one, all-vs-all). Denominator=NULL and Numerator=NULL selects all conditions and performs multiple all-vs-all comparisons. Log2FC values are obtained by dividing numerator by denominator, thus positive Log2FC values indicate higher expression in numerator. Default: c(conditions="Conditions", numerator=NULL, denumerator=NULL).
- pval
Character (optional). Abbreviation of the selected test to calculate p-value. For one-vs-one comparisons choose t.test, wilcox.test, chisq.test, cor.test, or lmFit (limma). For one-vs-all or all-vs-all comparisons choose aov (anova), welch (welch anova), kruskal.test, or lmFit (limma). Default: "lmFit".
- padj
Character (optional). Abbreviation of the selected p-value adjustment method for multiple hypothesis testing correction. See ?p.adjust for methods: "BH", "fdr", "bonferroni", "holm", etc. Default: "fdr".
- metadata_feature
Data frame (optional). Provides metadata information (e.g., pathway, retention time) for each metabolite. Only used if data is not a SummarizedExperiment. Row names must match metabolite names in data columns. Default: NULL.
- core
Logical (optional). Whether consumption/release input is used. Default: FALSE.
- vst
Logical. Whether to use variance stabilizing transformation on data when linear modeling is used for hypothesis testing. Default: FALSE.
- shapiro
Logical. Whether to perform Shapiro-Wilk test to assess data distribution (normal versus non-normal). Default: TRUE.
- bartlett
Logical. Whether to perform Bartlett's test. Default: TRUE.
- transform
Logical. If TRUE, data is expected to be non-log2-transformed and log2 transformation will be performed within limma and Log2FC calculation. If FALSE, data is expected to be log2-transformed as this impacts Log2FC calculation and limma. Default: TRUE.
- save_plot
Character (optional). File type of output plots: "svg", "png", "pdf". Default: "svg".
- save_table
Character (optional). File type for analysis results: "csv", "xlsx", "txt". Default: "csv".
- print_plot
Logical (optional). Whether volcano plot is printed as overview of results. Default: TRUE.
- path
Character (optional). Path to folder where results should be saved. Default: NULL.
Value
List of lists. Depending on parameter settings, returns dma (data frame of each comparison), shapiro (includes data frame and plot), bartlett (includes data frame and histogram), vst (includes data frame and plot), and VolcanoPlot (plots of each comparison).
Examples
data(intracell_raw_se)
ResI <- dma(
data = intracell_raw_se,
metadata_info = c(
Conditions = "Conditions", Numerator = NULL, Denominator = "HK2"
)
)
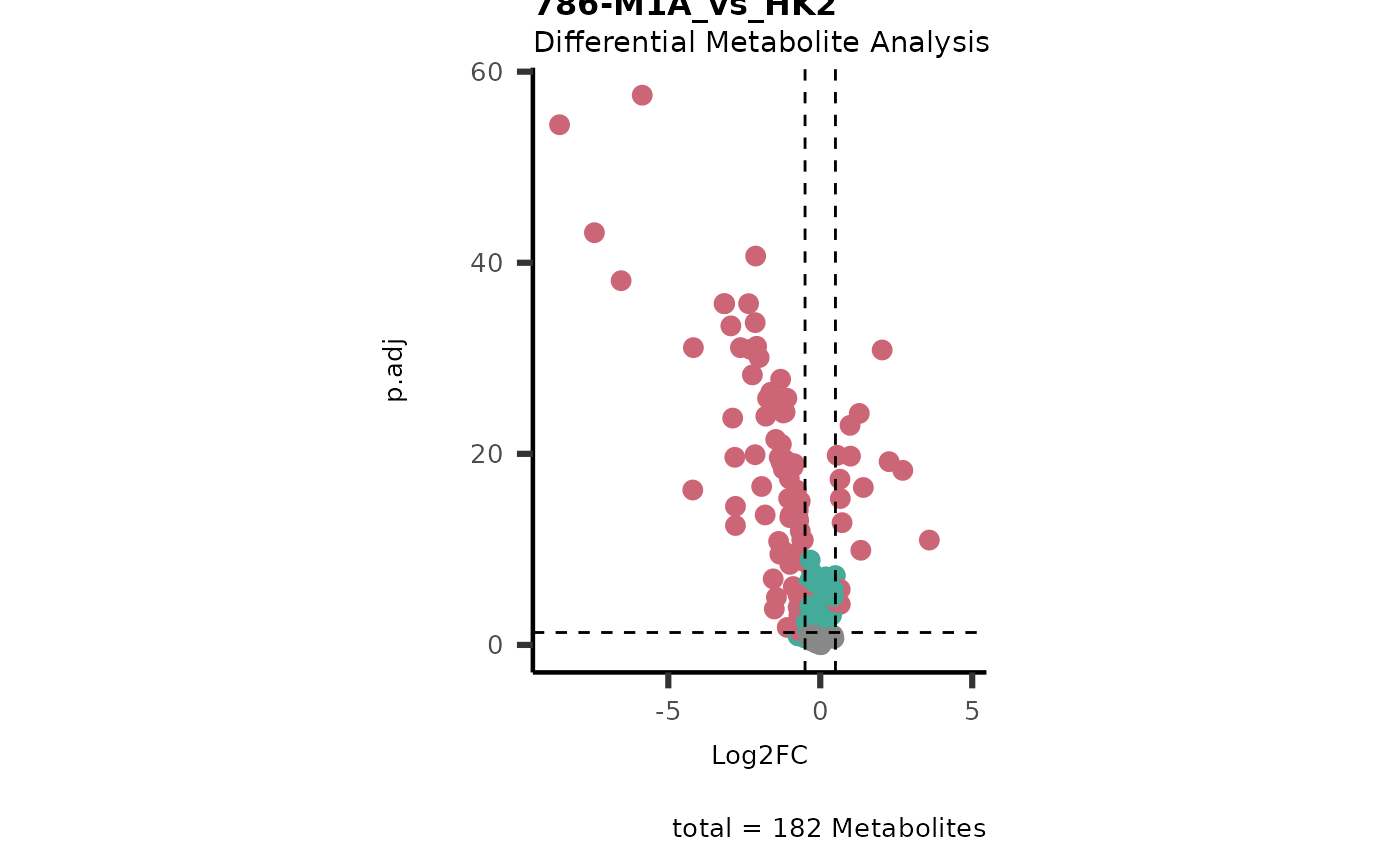
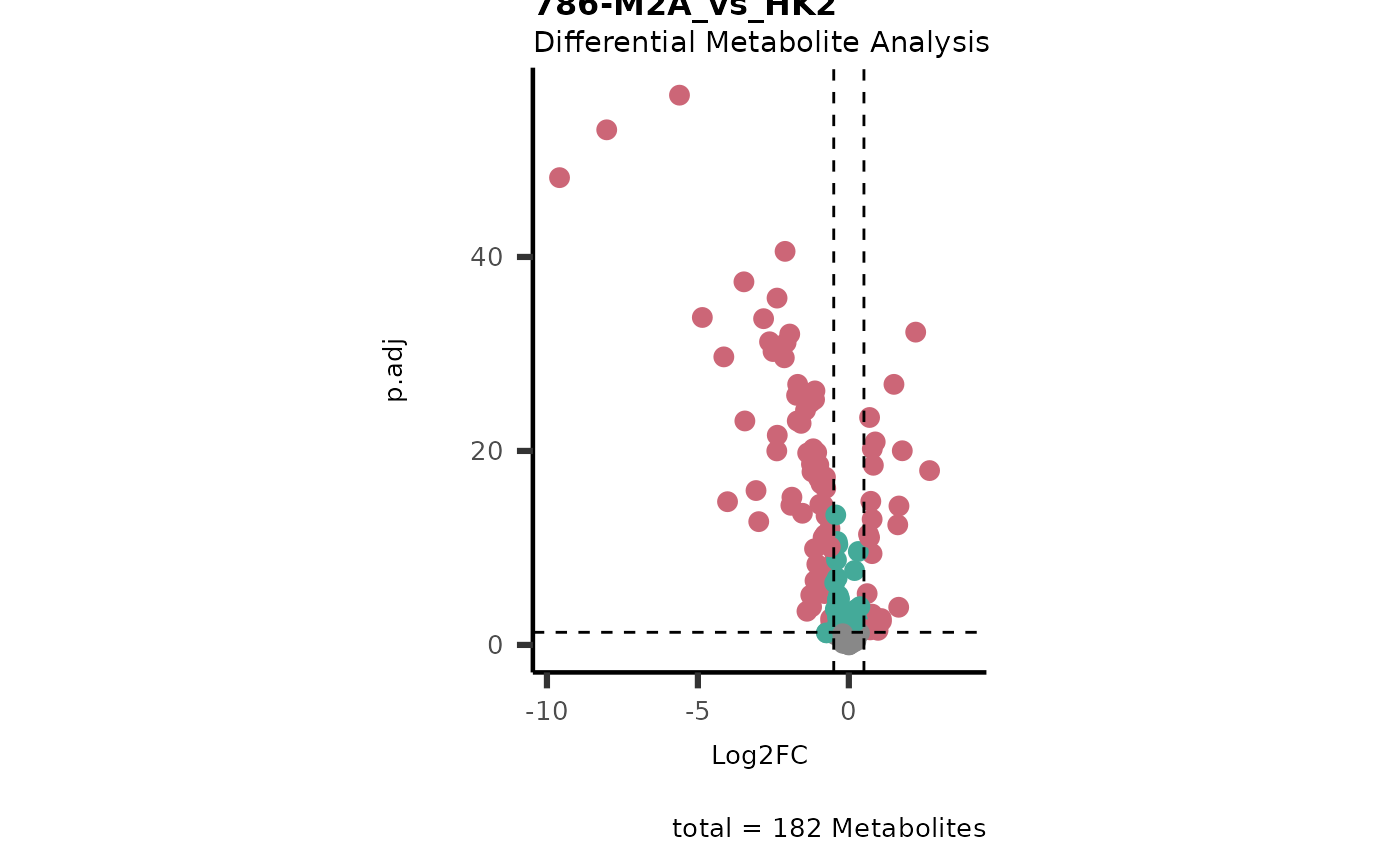
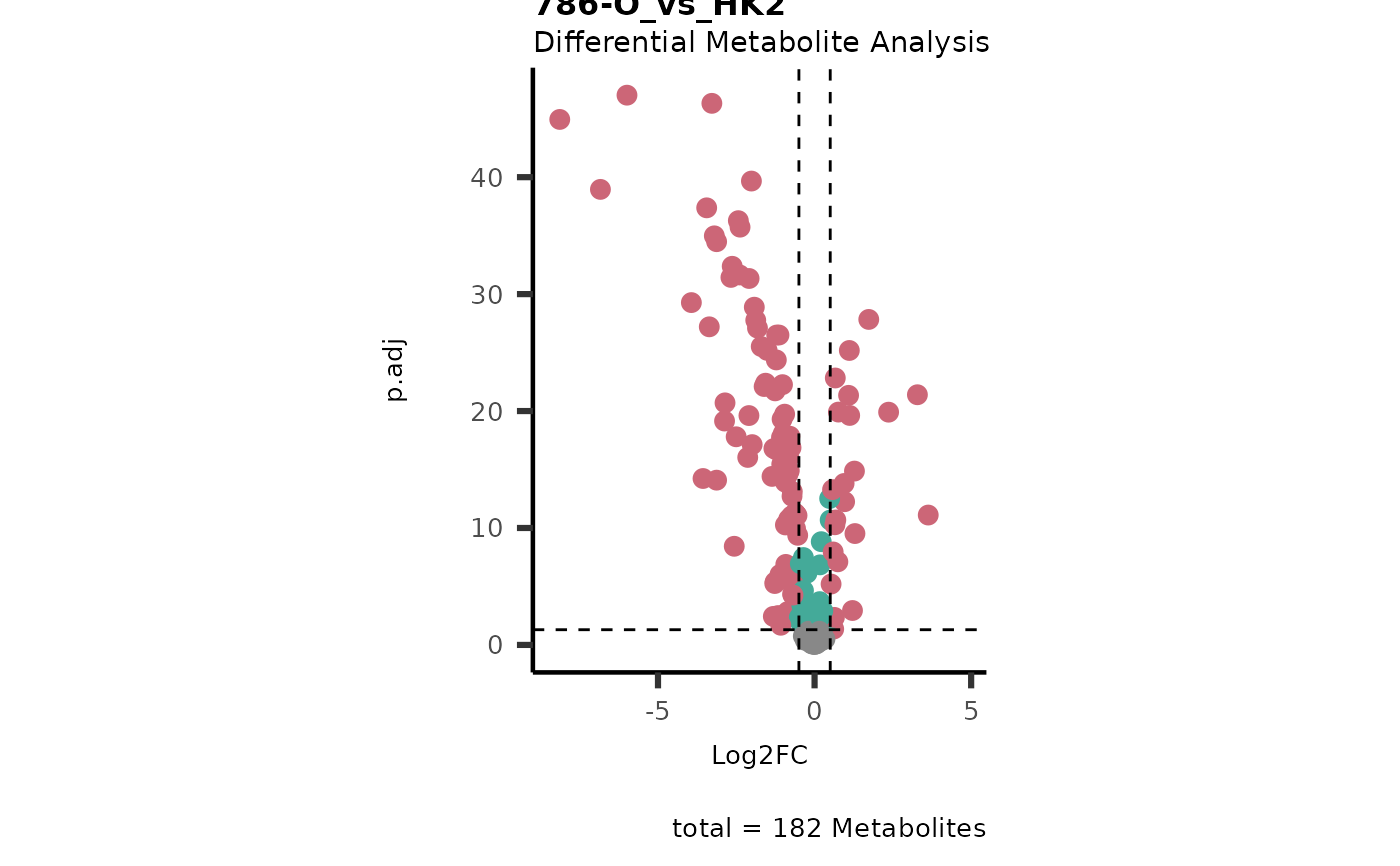
#> In `Numerator` 786-O, 786-M1A, 786-M2A, Pool, NA/0 values exist in 5 Metabolite(s). and in `denominator`HK2 2 Metabolite(s
#> ).. Those metabolite(s) might return p.val = NA, p.adj.= NA, t.val = NA. The Log2FC = Inf, if all replicates are 0/NA.
#> Warning: There are NA's / 0s in the data. This can impact the output of the SHapiro-Wilk test for all metabolites that include NAs / 0s.
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 527 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 527 rows containing non-finite outside the scale range
#> (`stat_density()`).
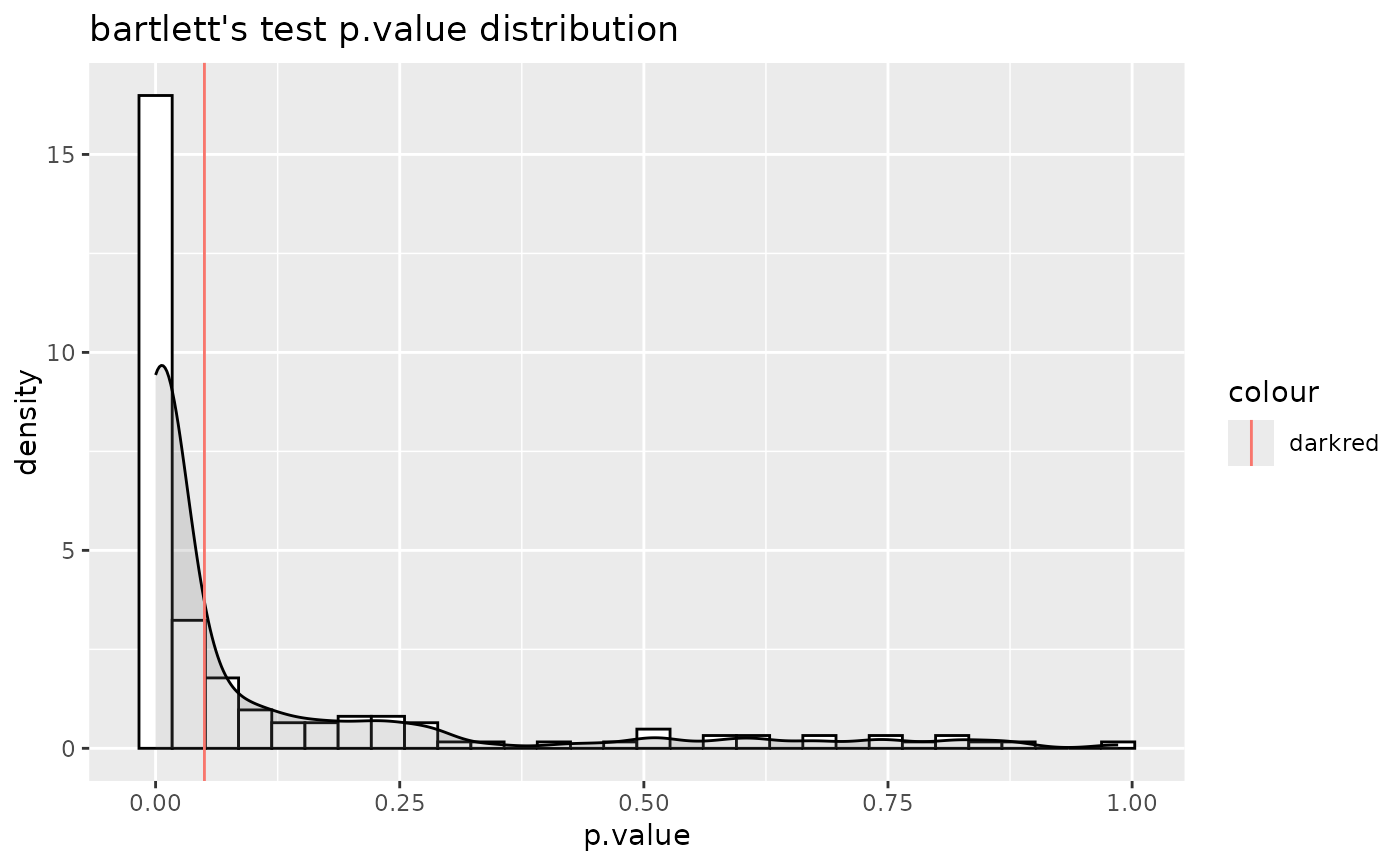
#> For 9.89% of metabolites the group variances are equal.
#> Warning: Partial NA coefficients for 1 probe(s)
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the EnhancedVolcano package.
#> Please report the issue to the authors.
#> Warning: The `size` argument of `element_line()` is deprecated as of ggplot2 3.4.0.
#> ℹ Please use the `linewidth` argument instead.
#> ℹ The deprecated feature was likely used in the EnhancedVolcano package.
#> Please report the issue to the authors.
#> Warning: Removed 556 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 556 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
 #> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
 #> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
 #> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 527 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 527 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 527 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 527 rows containing non-finite outside the scale range
#> (`stat_density()`).
 #> Warning: Removed 527 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 527 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: The dot-dot notation (`..density..`) was deprecated in ggplot2 3.4.0.
#> ℹ Please use `after_stat(density)` instead.
#> ℹ The deprecated feature was likely used in the MetaProViz package.
#> Please report the issue at <https://github.com/saezlab/MetaProViz/issues>.
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> Warning: Removed 527 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 527 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: The dot-dot notation (`..density..`) was deprecated in ggplot2 3.4.0.
#> ℹ Please use `after_stat(density)` instead.
#> ℹ The deprecated feature was likely used in the MetaProViz package.
#> Please report the issue at <https://github.com/saezlab/MetaProViz/issues>.
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
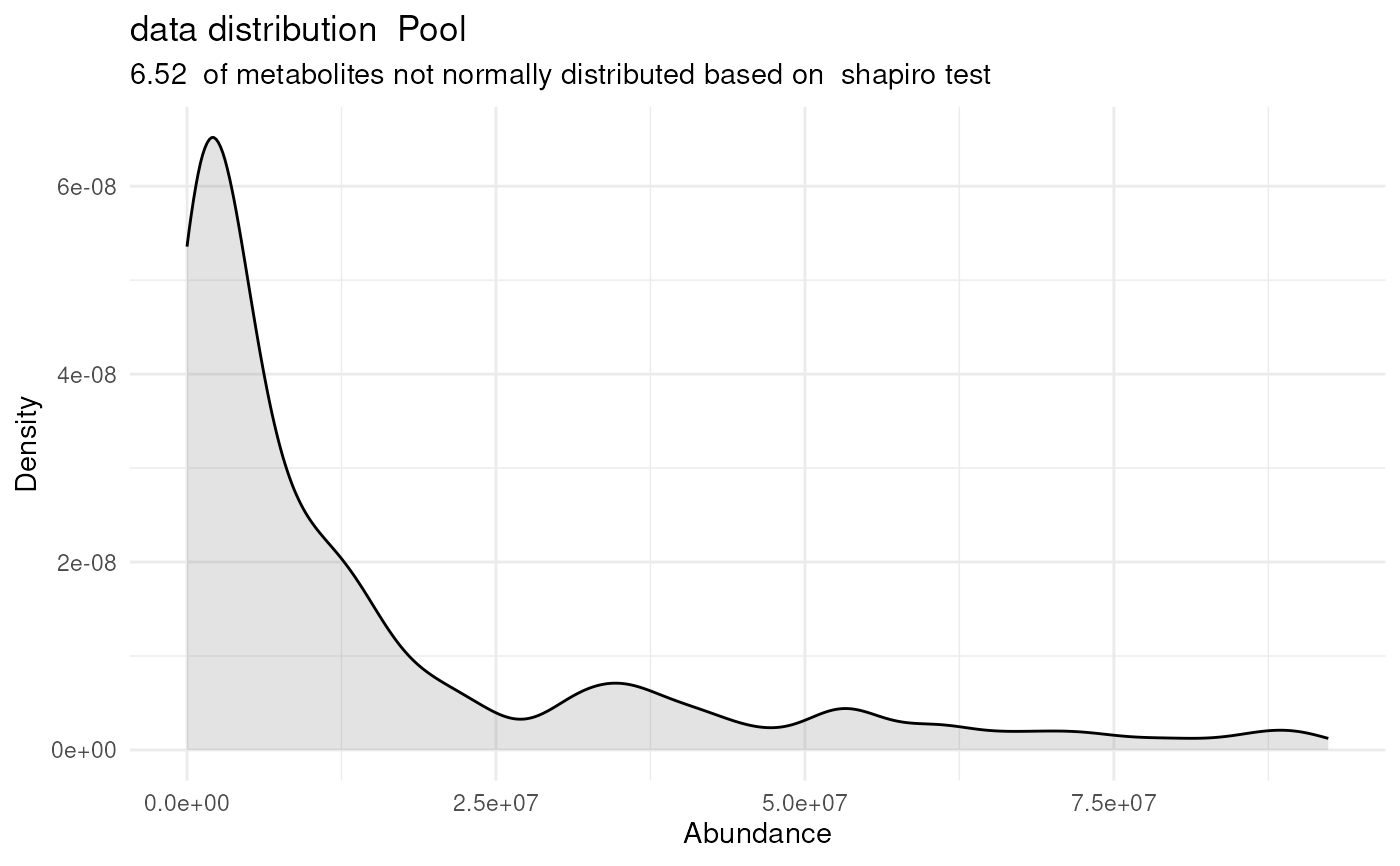 #> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
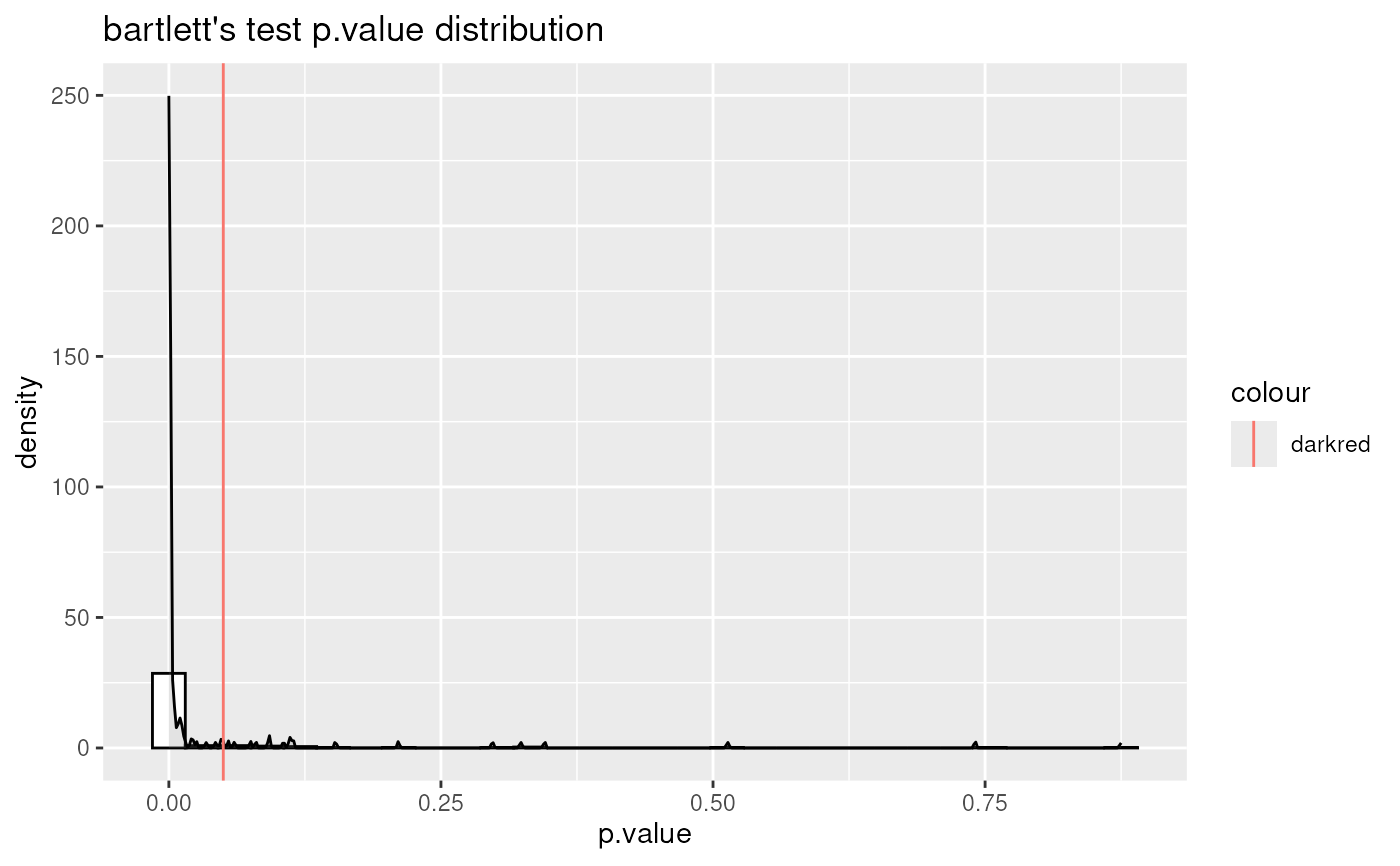
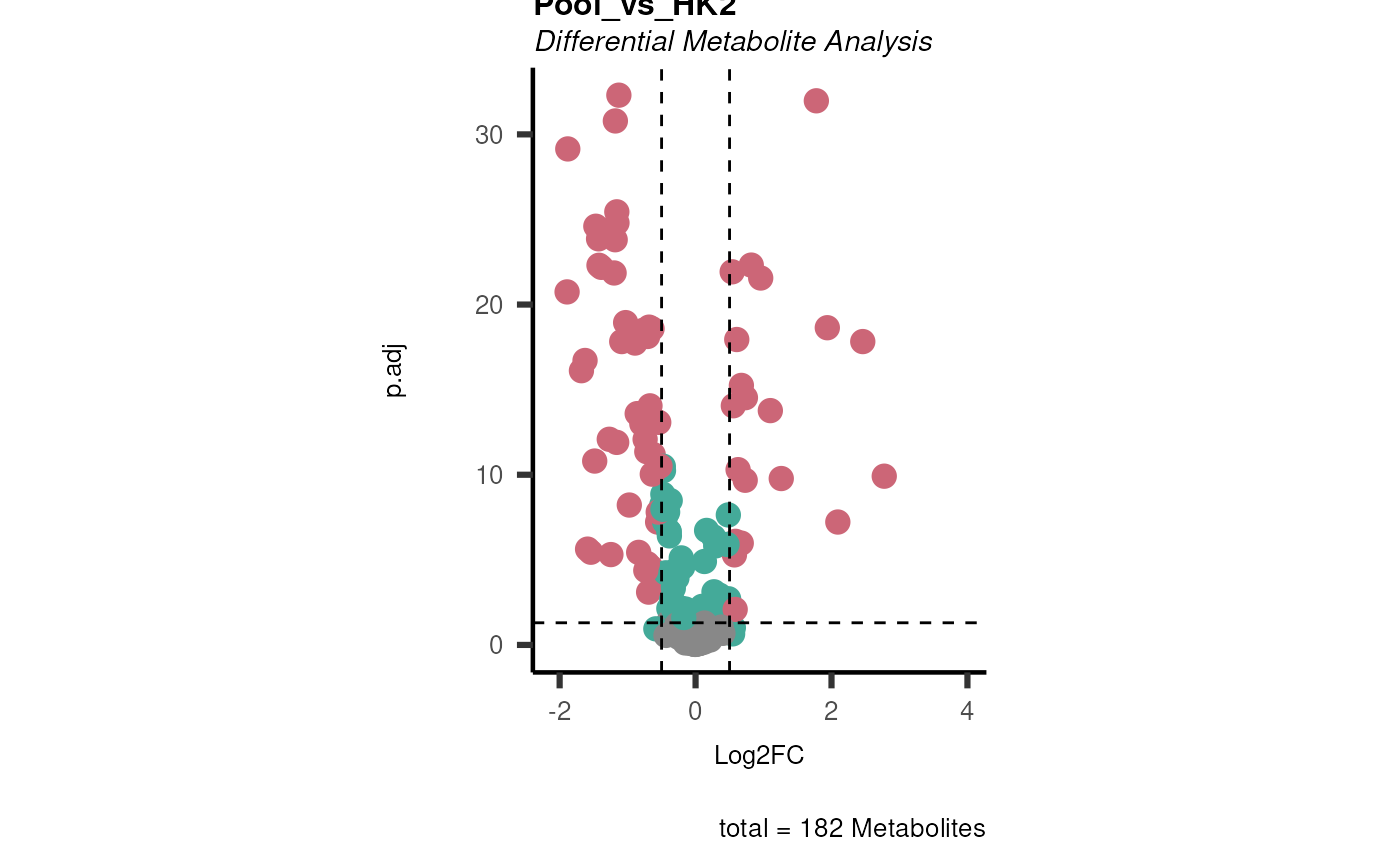
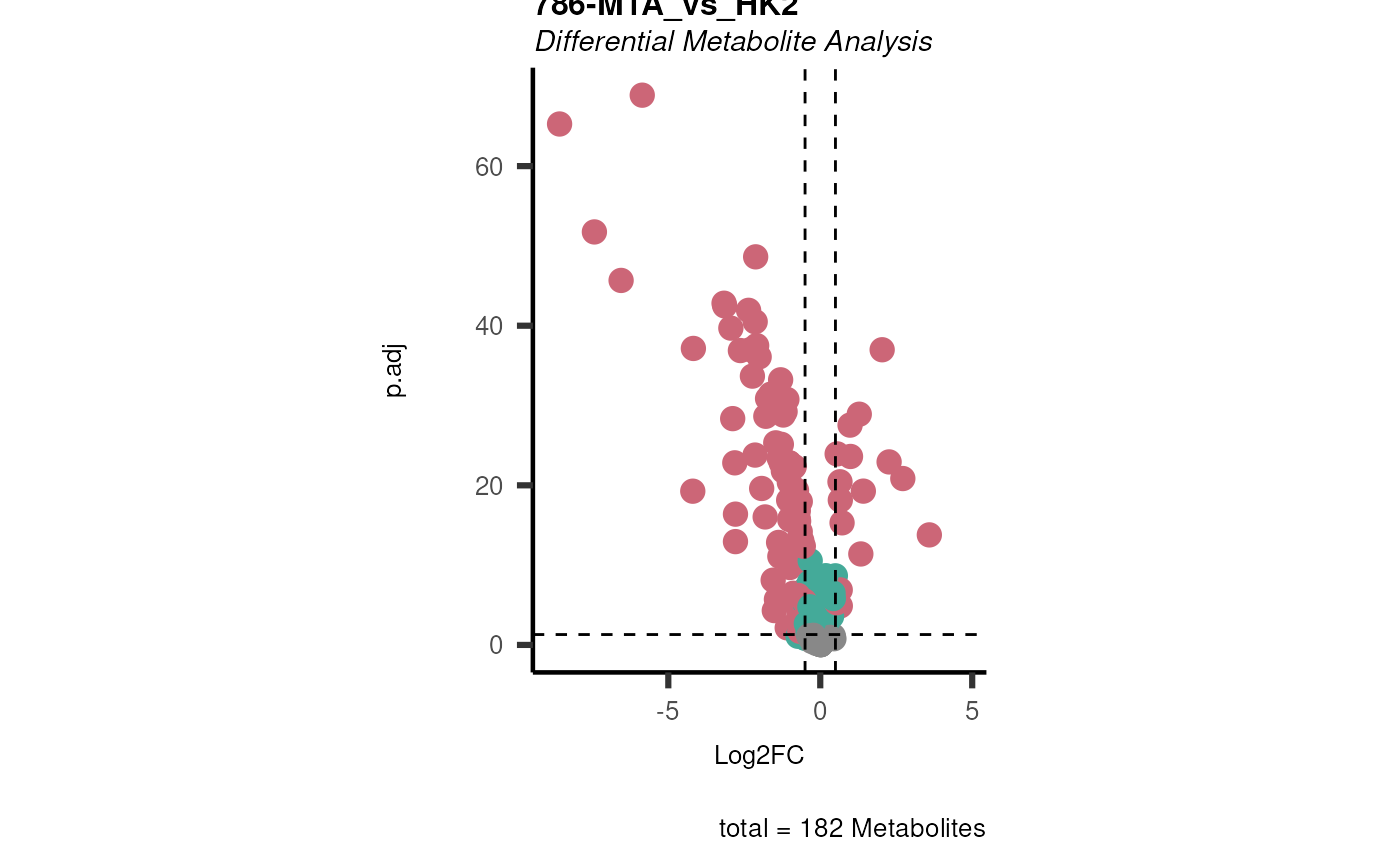
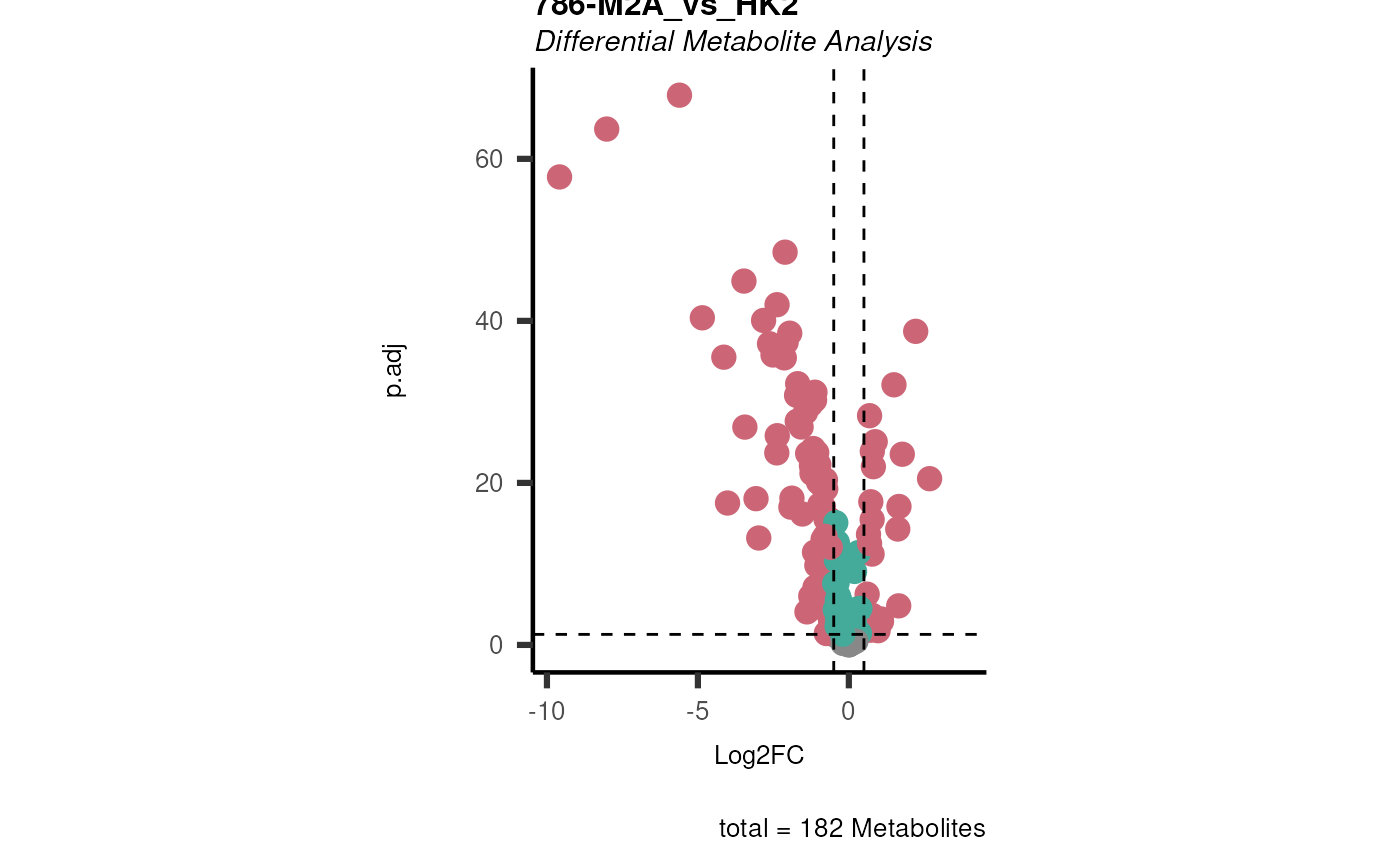
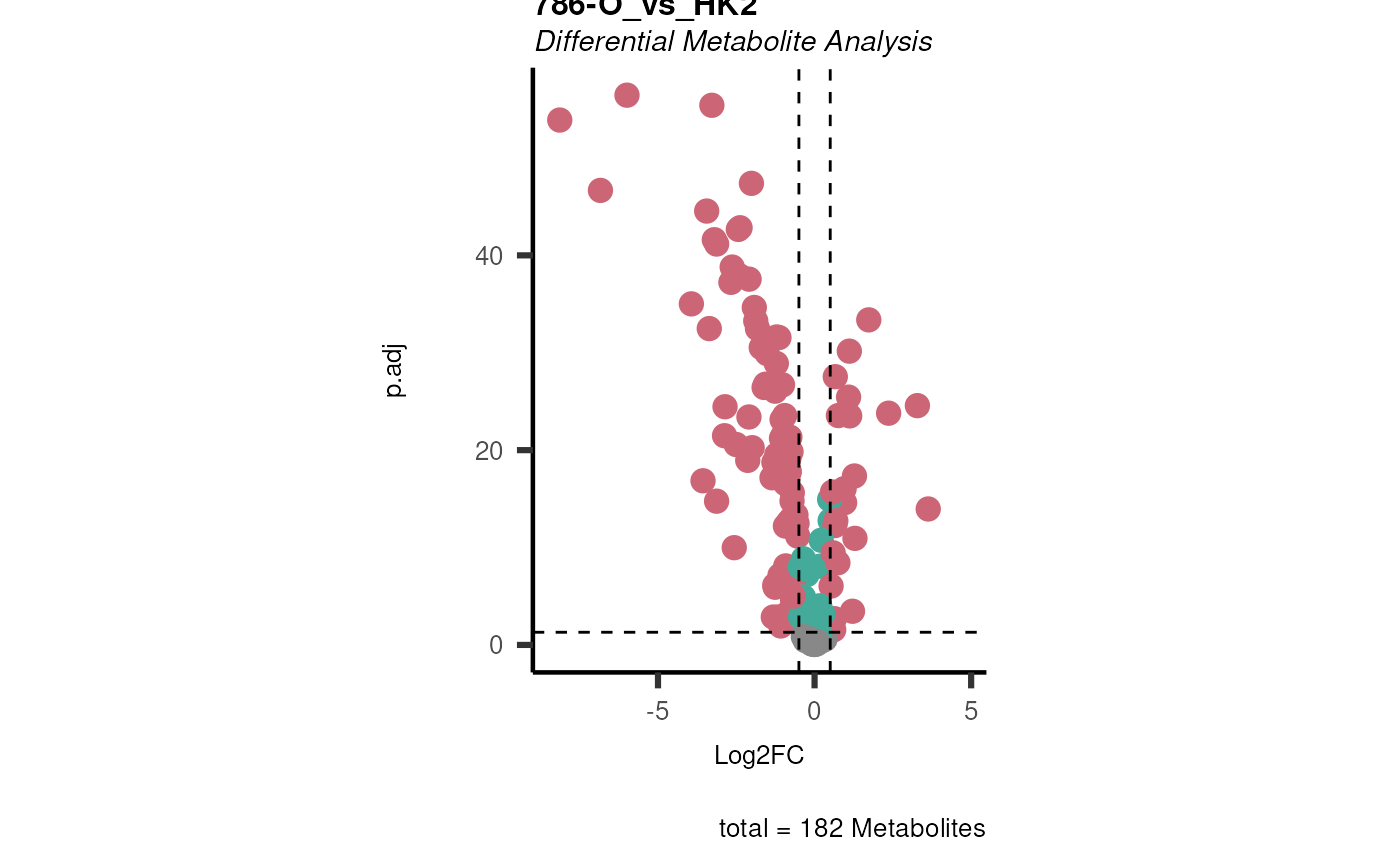 data(intracell_raw)
Intra <- intracell_raw[-c(49:58), ] %>% tibble::column_to_rownames("Code")
ResI <- dma(
data = Intra[, -c(1:3)],
metadata_sample = Intra[, c(1:3)],
metadata_info = c(
Conditions = "Conditions", Numerator = NULL, Denominator = "HK2"
)
)
#> In `Numerator` 786-O, 786-M1A, 786-M2A, NA/0 values exist in 5 Metabolite(s). and in `denominator`HK2 2 Metabolite(s
#> ).. Those metabolite(s) might return p.val = NA, p.adj.= NA, t.val = NA. The Log2FC = Inf, if all replicates are 0/NA.
#> Warning: There are NA's / 0s in the data. This can impact the output of the SHapiro-Wilk test for all metabolites that include NAs / 0s.
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
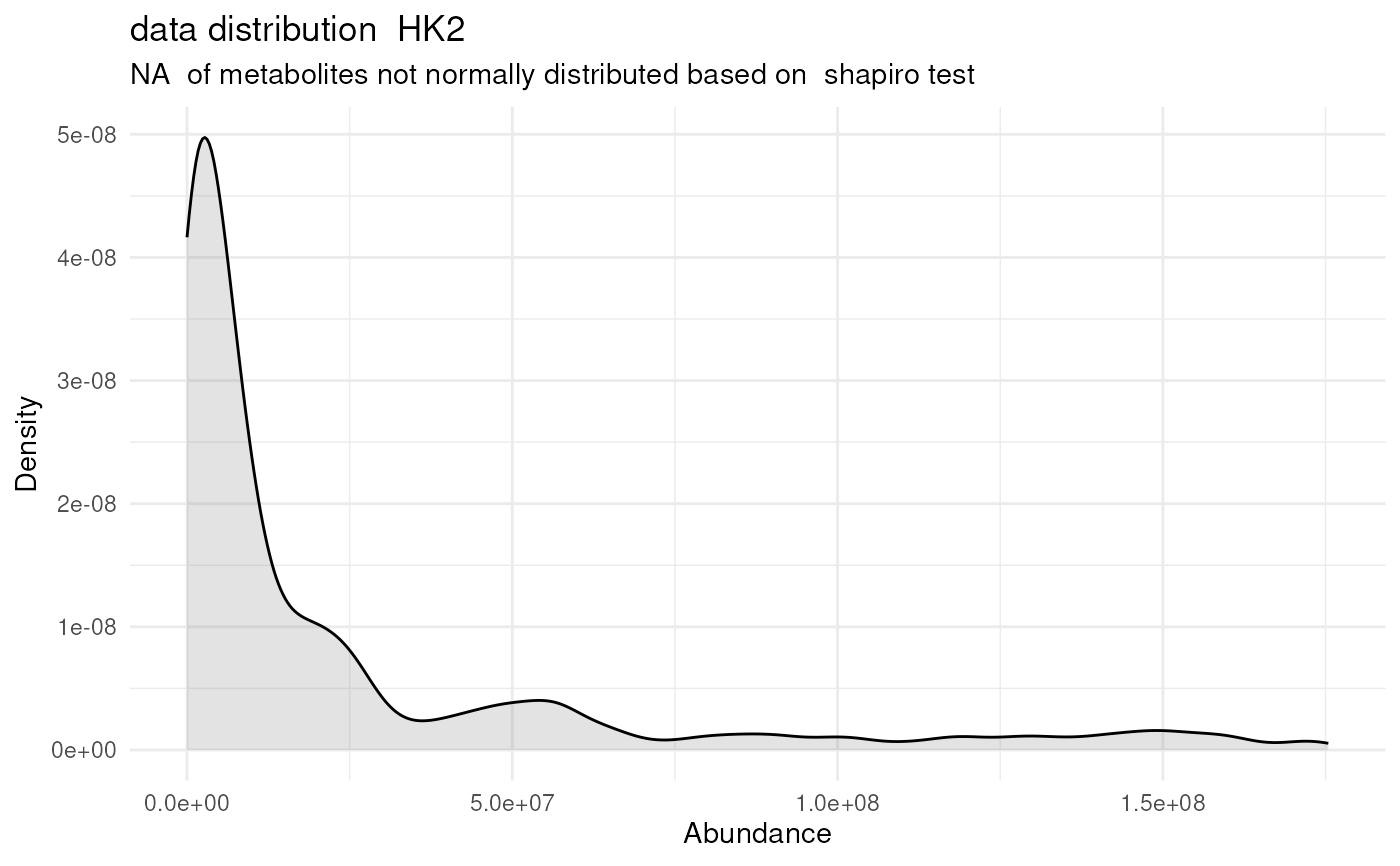
#> For 32.97% of metabolites the group variances are equal.
#> Warning: Partial NA coefficients for 1 probe(s)
#> Warning: Removed 556 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 556 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
data(intracell_raw)
Intra <- intracell_raw[-c(49:58), ] %>% tibble::column_to_rownames("Code")
ResI <- dma(
data = Intra[, -c(1:3)],
metadata_sample = Intra[, c(1:3)],
metadata_info = c(
Conditions = "Conditions", Numerator = NULL, Denominator = "HK2"
)
)
#> In `Numerator` 786-O, 786-M1A, 786-M2A, NA/0 values exist in 5 Metabolite(s). and in `denominator`HK2 2 Metabolite(s
#> ).. Those metabolite(s) might return p.val = NA, p.adj.= NA, t.val = NA. The Log2FC = Inf, if all replicates are 0/NA.
#> Warning: There are NA's / 0s in the data. This can impact the output of the SHapiro-Wilk test for all metabolites that include NAs / 0s.
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
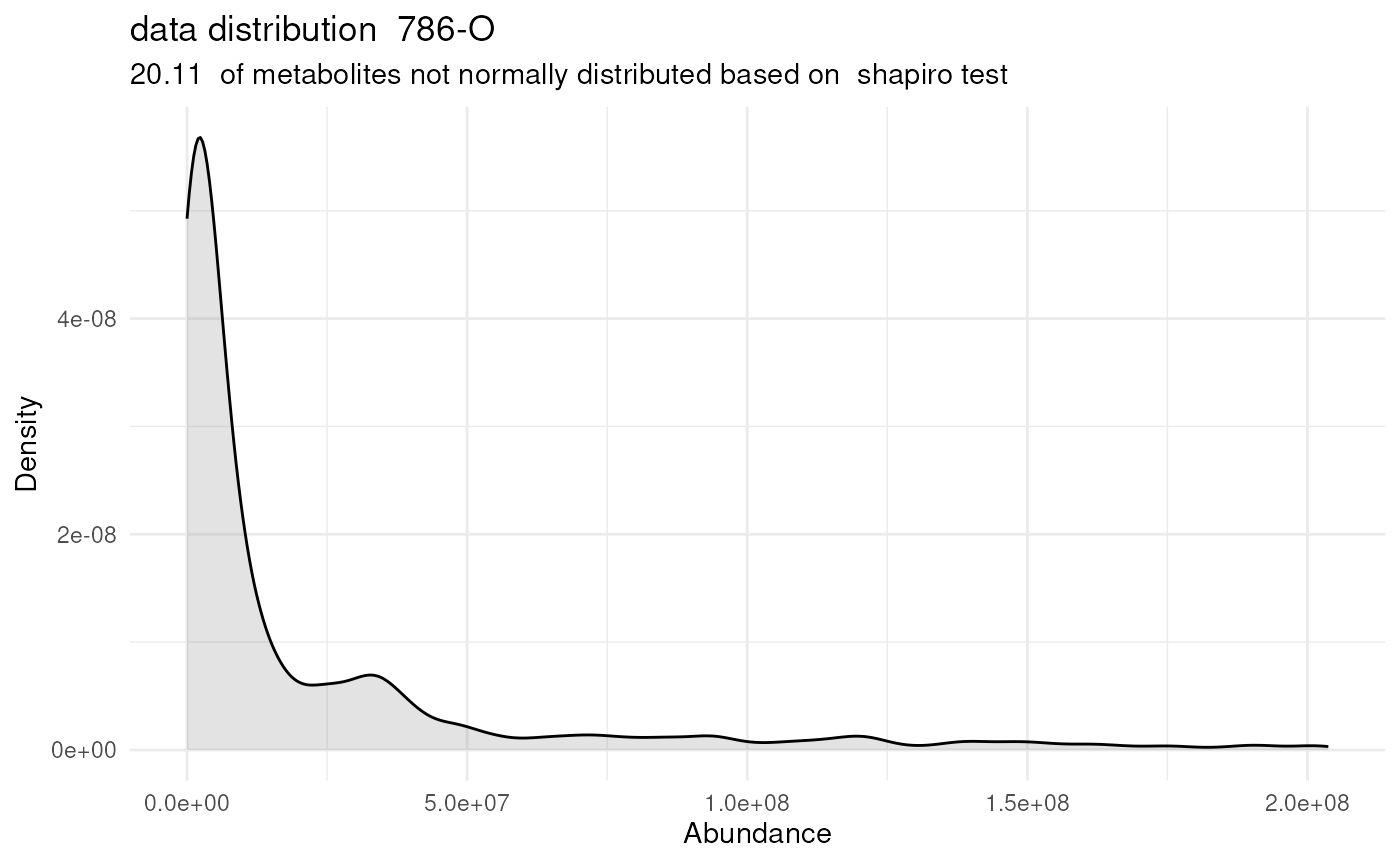
#> For 32.97% of metabolites the group variances are equal.
#> Warning: Partial NA coefficients for 1 probe(s)
#> Warning: Removed 556 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 556 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 556 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
 #> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 461 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 461 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
 #> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 458 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 458 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
 #> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> Warning: Removed 614 rows containing non-finite outside the scale range (`stat_bin()`).
#> Warning: Computation failed in `stat_bin()`.
#> Caused by error in `bin_breaks_width()`:
#> ! The number of histogram bins must be less than 1,000,000.
#> ℹ Did you make `binwidth` too small?
#> Warning: Removed 614 rows containing non-finite outside the scale range
#> (`stat_density()`).
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
 #> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.
#> `stat_bin()` using `bins = 30`. Pick better value `binwidth`.