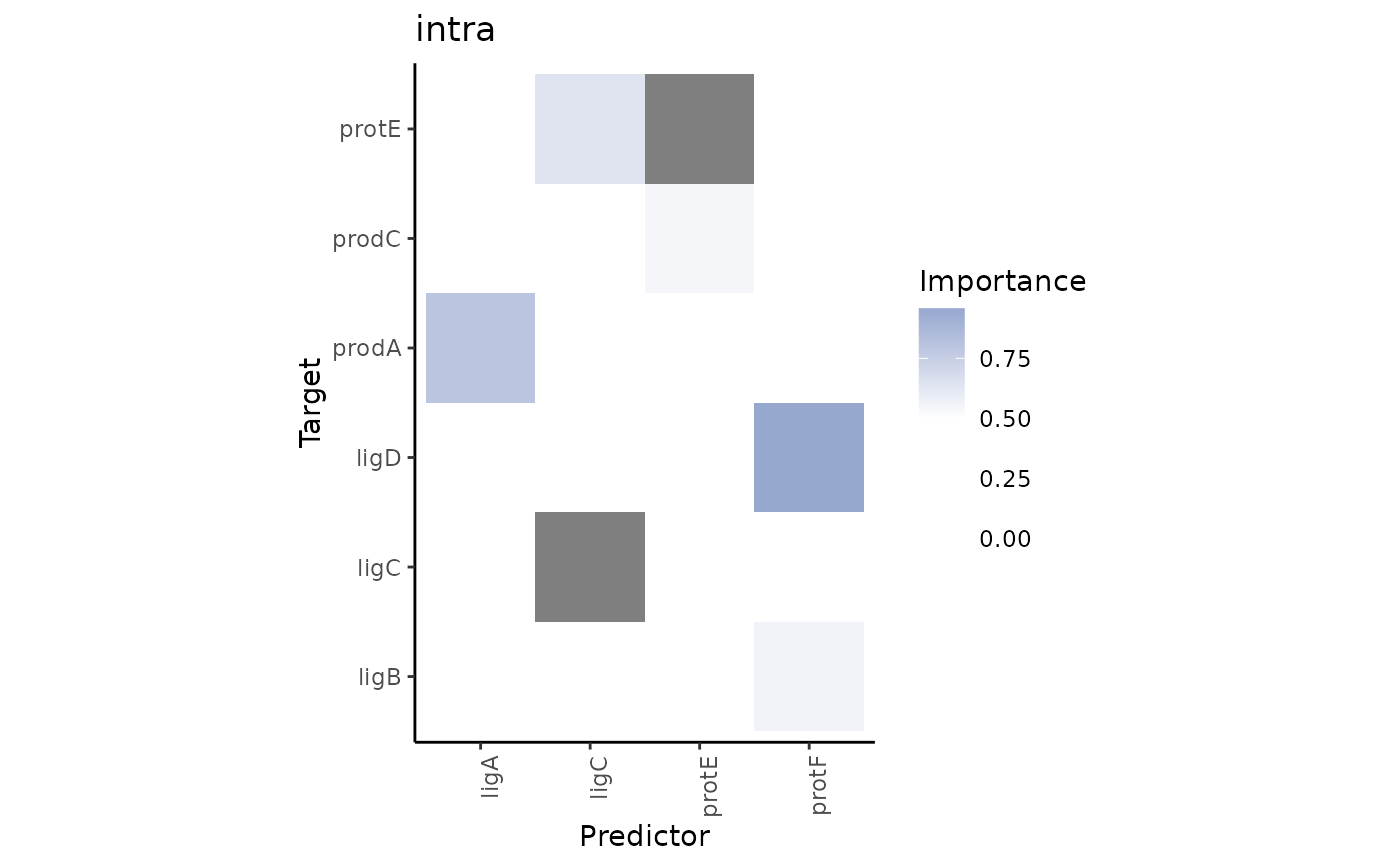
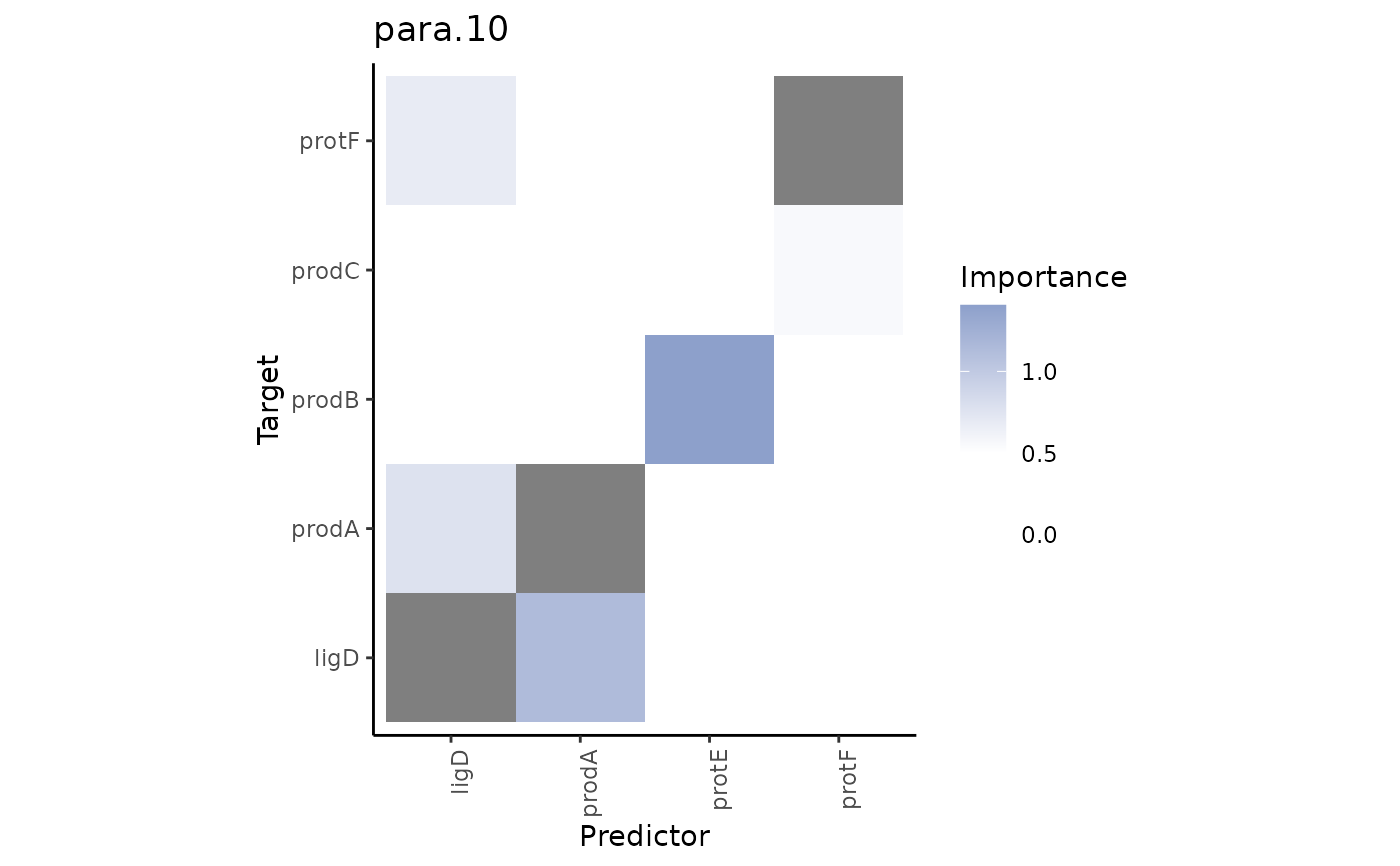
Plot interexperiment contrast of views.
Usage
plot_contrast_results(
misty.results.from,
misty.results.to,
views = NULL,
cutoff.from = 1,
cutoff.to = 1,
trim = -Inf,
trim.measure = c("gain.R2", "multi.R2", "intra.R2", "gain.RMSE", "multi.RMSE",
"intra.RMSE")
)Arguments
- misty.results.from, misty.results.to
a results list generated by
collect_results().- views
one or more abbreviated names of views.
- cutoff.from, cutoff.to
importance thresholds respective to the result lists.
- trim
display targets with performance value above (if R2 or gain) or below (otherwise) this value only.
- trim.measure
the measure used for trimming.
Details
The heatmaps show the interactions that are present and have importance above
a cutoff.to value in the views of misty.results.to but
not present or have importance below cutoff.from in the views
of misty.results.from.
See also
collect_results() to generate a
results list from raw results.
Other plotting functions:
plot_contrast_heatmap(),
plot_improvement_stats(),
plot_interaction_communities(),
plot_interaction_heatmap(),
plot_view_contributions()
Examples
# if for example the available samples come from different grades of tumors
grade1.results <- collect_results(c("results/synthetic1", "results/synthetic2"))
#>
#> Collecting improvements
#>
#> Collecting contributions
#>
#> Collecting importances
#>
#> Aggregating
grade3.results <- collect_results("results/synthetic10")
#>
#> Collecting improvements
#>
#> Collecting contributions
#>
#> Collecting importances
#>
#> Aggregating
# highlight interactions present in grade 1 tumors but not in grade 3 tumors
# in the paraview
grade3.results %>% plot_contrast_results(grade1.results, views = "para.10")
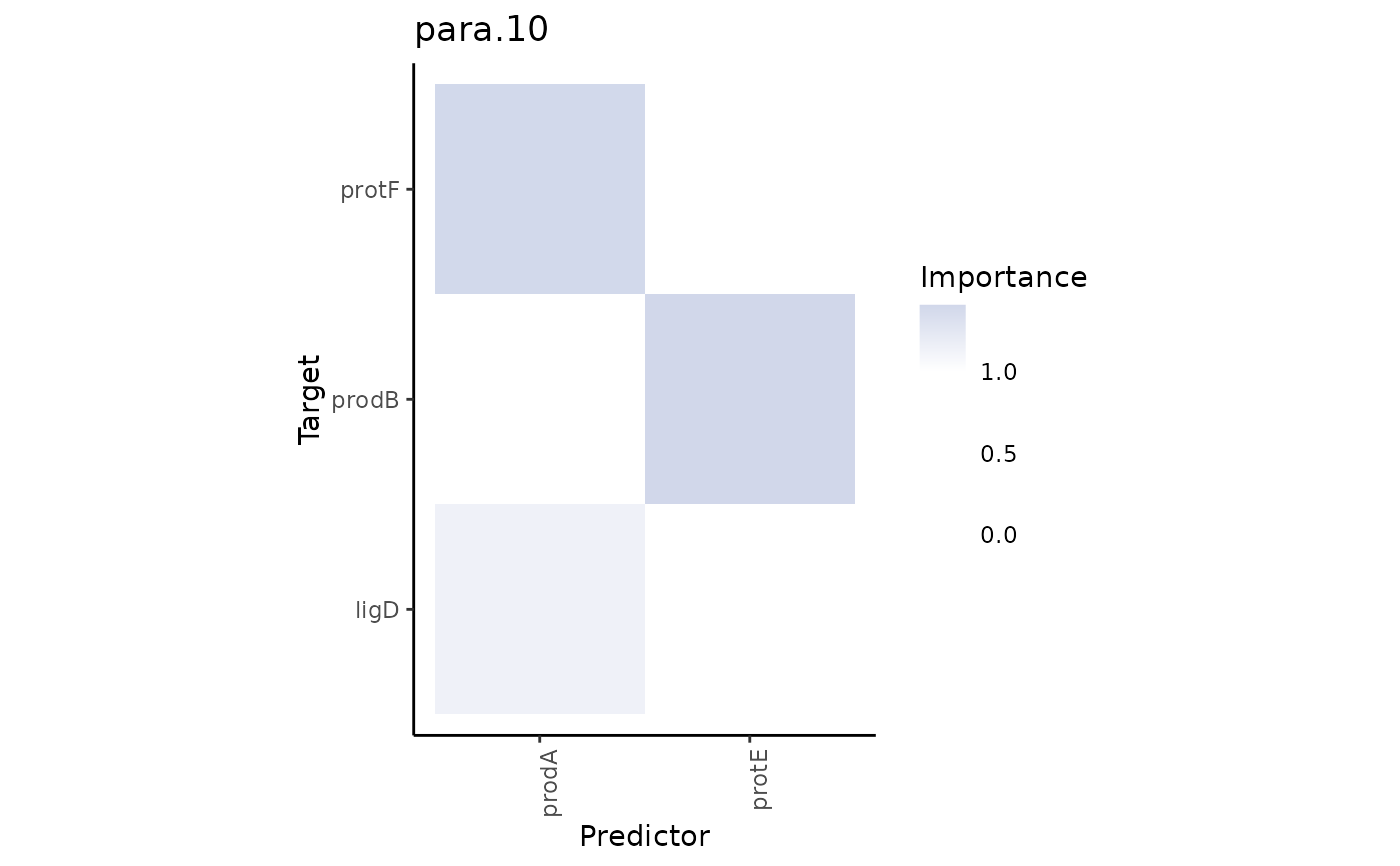 # see the loss of interactions in all views with lower sensitivity
plot_contrast_results(grade3.results, grade1.results, cutoff.from = 0.75, cutoff.to = 0.5)
# see the loss of interactions in all views with lower sensitivity
plot_contrast_results(grade3.results, grade1.results, cutoff.from = 0.75, cutoff.to = 0.5)